4UOQ
 
 | | Nucleophile mutant (E324A) of beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 | | Descriptor: | BETA-GALACTOSIDASE, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, ... | | Authors: | Viborg, A.H, Fredslund, F, Katayama, T, Nielsen, S.K, Svensson, B, Kitaoka, M, Lo Leggio, L, Abou Hachem, M. | | Deposit date: | 2014-06-09 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A beta 1-6/ beta 1-3 galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 gives insight into sub-specificities of beta-galactoside catabolism within Bifidobacterium.
Mol. Microbiol., 2014
|
|
4UOZ
 
 | | beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 nucleophile mutant E324A in complex with galactose | | Descriptor: | BETA-GALACTOSIDASE, TRIETHYLENE GLYCOL, ZINC ION, ... | | Authors: | Viborg, A.H, Fredslund, F, Katayama, T, Nielsen, S.K, Svensson, B, Kitaoka, M, Lo Leggio, L, Abou Hachem, M. | | Deposit date: | 2014-06-11 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A beta 1-6/ beta 1-3 galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 gives insight into sub-specificities of beta-galactoside catabolism within Bifidobacterium.
Mol. Microbiol., 2014
|
|
5T7K
 
 | | X-ray crystal structure of AA13 LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AoAA13, ZINC ION | | Authors: | Frandsen, K.E.H, Poulsen, J.-C.N, Tovborg, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2016-09-05 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Learning from oligosaccharide soaks of crystals of an AA13 lytic polysaccharide monooxygenase: crystal packing, ligand binding and active-site disorder.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5T7N
 
 | | X-ray crystal structure of AA13 LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AoAA13, ZINC ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.-C.N, Tovborg, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2016-09-05 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Learning from oligosaccharide soaks of crystals of an AA13 lytic polysaccharide monooxygenase: crystal packing, ligand binding and active-site disorder.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
8QCL
 
 | | A carbohydrate esterase family 15 (CE15) glucuronoyl esterase from Phocaeicola vulgatus ATCC 8482 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Banerjee, S, Poulsen, J.N, Mazurkewich, S, Seveso, A, Larsbrink, J, Lo Leggio, L. | | Deposit date: | 2023-08-27 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Polysaccharide utilization loci from Bacteroidota encode CE15 enzymes with possible roles in cleaving pectin-lignin bonds.
Appl.Environ.Microbiol., 90, 2024
|
|
5ACF
 
 | | X-ray Structure of LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.N, Tovborg, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2015-08-17 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular basis of polysaccharide cleavage by lytic polysaccharide monooxygenases.
Nat. Chem. Biol., 12, 2016
|
|
6Q5G
 
 | |
5ACH
 
 | | X-ray Structure of LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.N, Tovborg, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2015-08-17 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | The molecular basis of polysaccharide cleavage by lytic polysaccharide monooxygenases.
Nat. Chem. Biol., 12, 2016
|
|
1UT4
 
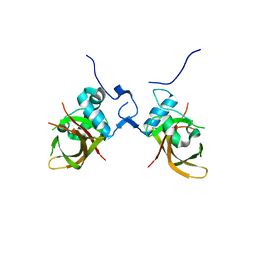 | | Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors | | Descriptor: | NO APICAL MERISTEM PROTEIN | | Authors: | Ernst, H.A, Olsen, A.N, Skriver, K, Larsen, S, Lo Leggio, L. | | Deposit date: | 2003-12-03 | | Release date: | 2004-03-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Conserved Domain of Anac, a Member of the Nac Family of Transcription Factors
Embo Rep., 5, 2004
|
|
1UT7
 
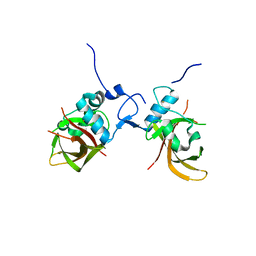 | | Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors | | Descriptor: | GOLD ION, NO APICAL MERISTEM PROTEIN | | Authors: | Ernst, H.A, Olsen, A.N, Skriver, K, Larsen, S, Lo Leggio, L. | | Deposit date: | 2003-12-04 | | Release date: | 2004-03-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Conserved Domain of Anac, a Member of the Nac Family of Transcription Factors
Embo Rep., 5, 2004
|
|
4UR4
 
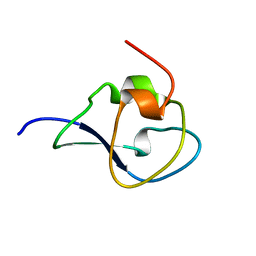 | | Structure of the type III fish antifreeze protein from Zoarces viviparus ZvAFP13 | | Descriptor: | ANTIFREEZE PROTEIN 13 | | Authors: | Wilkens, C, Poulsen, J.-C.N, Ramloev, H, Lo Leggio, L. | | Deposit date: | 2014-06-26 | | Release date: | 2014-07-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Purification, Crystal Structure Determination and Functional Characterization of Type III Antifreeze Proteins from the European Eelpout Zoarces Viviparus.
Cryobiology, 69, 2014
|
|
8QEF
 
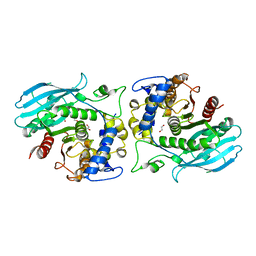 | | A carbohydrate esterase family 15 (CE15) glucuronoyl esterase from Phocaeicola ATCC 8482 bound to novel ligand. | | Descriptor: | 1,2-ETHANEDIOL, Putative acetyl xylan esterase, beta-D-galactopyranuronic acid | | Authors: | Banerjee, S, Poulsen, J.N, Mazurkewich, S, Seveso, A, Larsbrink, J, Lo Leggio, L. | | Deposit date: | 2023-08-31 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Polysaccharide utilization loci from Bacteroidota encode CE15 enzymes with possible roles in cleaving pectin-lignin bonds.
Appl.Environ.Microbiol., 90, 2024
|
|
8QAO
 
 | |
1R8L
 
 | | The structure of endo-beta-1,4-galactanase from Bacillus licheniformis | | Descriptor: | CALCIUM ION, endo-beta-1,4-galactanase | | Authors: | Ryttersgaard, C, Le Nours, J, Lo Leggio, L, Jorgensen, C.T, Christensen, L.L, Bjornvad, M, Larsen, S. | | Deposit date: | 2003-10-27 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products
J.Mol.Biol., 341, 2004
|
|
3EII
 
 | | Zinc-bound glycoside hydrolase 61 E from Thielavia terrestris | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Salbo, R, Welner, D, Lo Leggio, L, Harris, P, McFarland, K. | | Deposit date: | 2008-09-16 | | Release date: | 2009-10-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family.
Biochemistry, 49, 2010
|
|
3EJA
 
 | | Magnesium-bound glycoside hydrolase 61 isoform E from Thielavia terrestris | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Salbo, R, Welner, D, Lo Leggio, L, Harris, P, McFarland, K. | | Deposit date: | 2008-09-18 | | Release date: | 2009-10-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family.
Biochemistry, 49, 2010
|
|
1UR4
 
 | | The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products. | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GALACTANASE, ... | | Authors: | Ryttersgaard, C, Le Nours, J, Lo Leggio, L, Jorgensen, C.T, Christensen, L.L.H, Bjornvad, M, Larsen, S. | | Deposit date: | 2003-10-24 | | Release date: | 2004-10-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of Endo-Beta-1,4-Galactanase from Bacillus Licheniformis in Complex with Two Oligosaccharide Products
J.Mol.Biol., 341, 2004
|
|
1UR0
 
 | | The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products. | | Descriptor: | CALCIUM ION, GALACTANASE, TRIETHYLENE GLYCOL, ... | | Authors: | Ryttersgaard, C, Le Nours, J, Lo Leggio, L, Jorgensen, C.T, Christensen, L.L.H, Bjornvad, M, Larsen, S. | | Deposit date: | 2003-10-24 | | Release date: | 2004-10-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structure of Endo-Beta-1,4-Galactanase from Bacillus Licheniformis in Complex with Two Oligosaccharide Products
J.Mol.Biol., 341, 2004
|
|
3NJV
 
 | | Rhamnogalacturonan lyase from Aspergillus aculeatus K150A substrate complex | | Descriptor: | CALCIUM ION, Rhamnogalacturonase B, SULFATE ION, ... | | Authors: | Jensen, M.H, Otten, H, Christensen, U, Borchert, T.V, Christensen, L.L.H, Larsen, S, Lo Leggio, L. | | Deposit date: | 2010-06-18 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Biochemical Studies Elucidate the Mechanism of Rhamnogalacturonan Lyase from Aspergillus aculeatus.
J.Mol.Biol., 404, 2010
|
|
4AIE
 
 | | Structure of glucan-1,6-alpha-glucosidase from Lactobacillus acidophilus NCFM | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLUCAN 1,6-ALPHA-GLUCOSIDASE, ... | | Authors: | Fredslund, F, Navarro Poulsen, J.C, Lo Leggio, L. | | Deposit date: | 2012-02-09 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Enzymology and Structure of the Gh13_31 Glucan 1,6-Alpha-Glucosidase that Confers Isomaltooligosaccharide Utilization in the Probiotic Lactobacillus Acidophilus Ncfm.
J.Bacteriol., 194, 2012
|
|
2XN2
 
 | | Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with galactose | | Descriptor: | ALPHA-GALACTOSIDASE, GLYCEROL, IMIDAZOLE, ... | | Authors: | Fredslund, F, Abou Hachem, M, Larsen, R.J, Sorensen, P.G, Lo Leggio, L, Svensson, B. | | Deposit date: | 2010-07-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of Alpha-Galactosidase from Lactobacillus Acidophilus Ncfm: Insight Into Tetramer Formation and Substrate Binding.
J.Mol.Biol., 412, 2011
|
|
2XN1
 
 | | Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALPHA-GALACTOSIDASE, GLYCEROL | | Authors: | Fredslund, F, Abou Hachem, M, Larsen, R.J, Sorensen, P.G, Lo Leggio, L, Svensson, B. | | Deposit date: | 2010-07-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Alpha-Galactosidase from Lactobacillus Acidophilus Ncfm: Insight Into Tetramer Formation and Substrate Binding.
J.Mol.Biol., 412, 2011
|
|
2XN0
 
 | | Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM, PtCl4 derivative | | Descriptor: | ALPHA-GALACTOSIDASE, GLYCEROL, PLATINUM (II) ION | | Authors: | Fredslund, F, Abou Hachem, M, Larsen, R.J, Sorensen, P.G, Lo Leggio, L, Svensson, B. | | Deposit date: | 2010-07-30 | | Release date: | 2011-08-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Alpha-Galactosidase from Lactobacillus Acidophilus Ncfm: Insight Into Tetramer Formation and Substrate Binding.
J.Mol.Biol., 412, 2011
|
|
7OS9
 
 | | Crystal Structure of Domain Swapped Trp Repressor V58I Variant with purification tag | | Descriptor: | IMIDAZOLE, Trp operon repressor | | Authors: | Sprenger, J, Lawson, C.L, Lo Leggio, L, Von Wachenfeldt, C, Carey, J. | | Deposit date: | 2021-06-08 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structures of Val58Ile tryptophan repressor in a domain-swapped array in the presence and absence of L-tryptophan.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
2X2Y
 
 | | Cellulomonas fimi endo-beta-1,4-mannanase double mutant | | Descriptor: | FORMIC ACID, MAGNESIUM ION, MAN26A | | Authors: | Hekmat, O, Lo Leggio, L, Rosengren, A, Kamarauskaite, J, Kolenova, K, Staalbrand, H. | | Deposit date: | 2010-01-18 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Rational Engineering of Mannosyl Binding in the Distal Glycone Subsites of Cellulomonas Fimi Endo-Beta-1,4-Mannanase: Mannosyl Binding Promoted at Subsite -2 and Demoted at Subsite -3 .
Biochemistry, 49, 2010
|
|
