5YAX
 
 | | Crystal structure of a human neutralizing antibody bound to a HBV preS1 peptide | | Descriptor: | Large envelope protein, SODIUM ION, scFv1 antibody | | Authors: | Liu, X, Zheng, S, Ye, K, Sui, J. | | Deposit date: | 2017-09-02 | | Release date: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A potent human neutralizing antibody Fc-dependently reduces established HBV infections
Elife, 6, 2017
|
|
7EE5
 
 | | Crystal structure of Neu5Gc bound PltC | | Descriptor: | N-glycolyl-alpha-neuraminic acid, N-glycolyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, Subtilase cytotoxin subunit B-like protein, ... | | Authors: | Liu, X.Y, Chen, Z, Gao, X. | | Deposit date: | 2021-03-17 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Molecular Insights into the Assembly and Functional Diversification of Typhoid Toxin.
Mbio, 13, 2022
|
|
7EE4
 
 | | Crystal structure of Neu5Ac bound PltC | | Descriptor: | N-acetyl-alpha-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Liu, X.Y, Chen, Z, Gao, X. | | Deposit date: | 2021-03-17 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Insights into the Assembly and Functional Diversification of Typhoid Toxin.
Mbio, 13, 2022
|
|
7EE3
 
 | | Crystal structure of PltC | | Descriptor: | Subtilase cytotoxin subunit B-like protein, TETRAETHYLENE GLYCOL | | Authors: | Liu, X.Y, Chen, Z, Gao, X. | | Deposit date: | 2021-03-17 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Molecular Insights into the Assembly and Functional Diversification of Typhoid Toxin.
Mbio, 13, 2022
|
|
2PKY
 
 | | The Effect of Deuteration on Protein Structure A High Resolution Comparison of Hydrogenous and Perdeuterated Haloalkane Dehalogenase | | Descriptor: | Haloalkane dehalogenase | | Authors: | Liu, X, Hanson, L, Langan, P, Viola, R.E. | | Deposit date: | 2007-04-18 | | Release date: | 2007-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The effect of deuteration on protein structure: a high-resolution comparison of hydrogenous and perdeuterated haloalkane dehalogenase.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
6AKI
 
 | | Calcium release-activated calcium channel protein 1, P288L mutant | | Descriptor: | CHLORIDE ION, Calcium release-activated calcium channel protein 1 | | Authors: | Liu, X, Wu, G, Yang, X, Shen, Y. | | Deposit date: | 2018-09-01 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (4.496 Å) | | Cite: | Calcium release-activated calcium channel protein 1, P288L mutant
To Be Published
|
|
4GVC
 
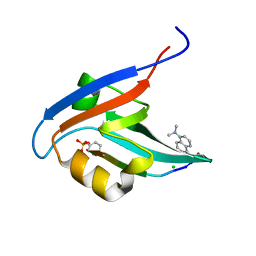 | | Crystal Structure of T-cell Lymphoma Invasion and Metastasis-1 PDZ in complex with phosphorylated Syndecan1 Peptide | | Descriptor: | 5-(DIMETHYLAMINO)-1-NAPHTHALENESULFONIC ACID(DANSYL ACID), CHLORIDE ION, SODIUM ION, ... | | Authors: | Liu, X, Shepherd, T.R, Murray, A.M, Xu, Z, Fuentes, E.J. | | Deposit date: | 2012-08-30 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The structure of the Tiam1 PDZ domain/ phospho-syndecan1 complex reveals a ligand conformation that modulates protein dynamics.
Structure, 21, 2013
|
|
4GQ1
 
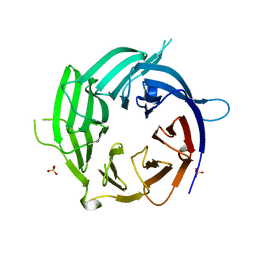 | | Nup37 of S. pombe | | Descriptor: | Nup37, SULFATE ION | | Authors: | Liu, X, Mitchell, J, Wozniak, R, Blobel, G, Fan, J. | | Deposit date: | 2012-08-22 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural evolution of the membrane-coating module of the nuclear pore complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GVD
 
 | | Crystal Structure of T-cell Lymphoma Invasion and Metastasis-1 PDZ in complex with Syndecan1 Peptide | | Descriptor: | 5-(DIMETHYLAMINO)-1-NAPHTHALENESULFONIC ACID(DANSYL ACID), CHLORIDE ION, SODIUM ION, ... | | Authors: | Liu, X, Shepherd, T.R, Murray, A.M, Xu, Z, Fuentes, E.J. | | Deposit date: | 2012-08-30 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of the Tiam1 PDZ domain/ phospho-syndecan1 complex reveals a ligand conformation that modulates protein dynamics.
Structure, 21, 2013
|
|
4GQ2
 
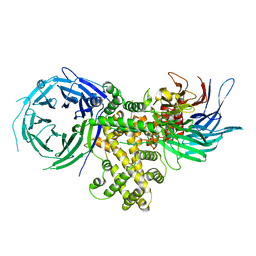 | | S. pombe Nup120-Nup37 complex | | Descriptor: | Nucleoporin nup120, Nup37 | | Authors: | Liu, X, Mitchell, J, Wozniak, R, Blobel, G, Fan, J. | | Deposit date: | 2012-08-22 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural evolution of the membrane-coating module of the nuclear pore complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7EE6
 
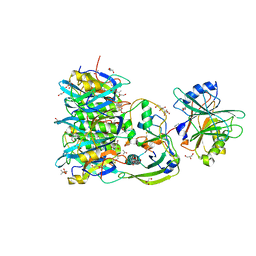 | | Crystal structure of PltC toxin | | Descriptor: | ACETONE, CITRATE ANION, Cytolethal distending toxin subunit B family protein, ... | | Authors: | Liu, X.Y, Chen, Z, Gao, X. | | Deposit date: | 2021-03-17 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Molecular Insights into the Assembly and Functional Diversification of Typhoid Toxin.
Mbio, 13, 2022
|
|
4KC3
 
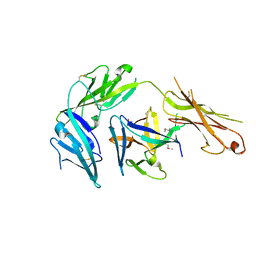 | | Cytokine/receptor binary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 receptor-like 1, Interleukin-33 | | Authors: | Liu, X, Wang, X.Q. | | Deposit date: | 2013-04-24 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.2702 Å) | | Cite: | Structural insights into the interaction of IL-33 with its receptors.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3R24
 
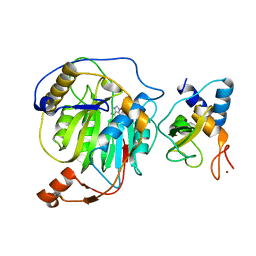 | | Crystal structure of nsp10/nsp16 complex of SARS coronavirus | | Descriptor: | 2'-O-methyl transferase, Non-structural protein 10 and Non-structural protein 11, S-ADENOSYLMETHIONINE, ... | | Authors: | Liu, X, Guo, D, Su, C, Chen, Y. | | Deposit date: | 2011-03-13 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex.
Plos Pathog., 7, 2011
|
|
4KFV
 
 | |
7E5X
 
 | |
5Y0I
 
 | | Solution structure of arenicin-3 derivative N1 | | Descriptor: | NZ17074(N1) | | Authors: | Liu, X.H, Wang, J.H. | | Deposit date: | 2017-07-17 | | Release date: | 2017-07-26 | | Method: | SOLUTION NMR | | Cite: | Antibacterial and detoxifying activity of NZ17074 analogues with multi-layers of selective antimicrobial actions against Escherichia coli and Salmonella enteritidis
Sci Rep, 7, 2017
|
|
5Y0H
 
 | | Solution structure of arenicin-3 derivative N6 | | Descriptor: | N6 | | Authors: | Liu, X.H, Wang, J.H. | | Deposit date: | 2017-07-17 | | Release date: | 2017-07-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Antibacterial and detoxifying activity of NZ17074 analogues with multi-layers of selective antimicrobial actions against Escherichia coli and Salmonella enteritidis
Sci Rep, 7, 2017
|
|
5Y0J
 
 | | Solution structure of arenicin-3 derivative N2 | | Descriptor: | N2 | | Authors: | Liu, X.H, Wang, J.H. | | Deposit date: | 2017-07-17 | | Release date: | 2017-07-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Antibacterial and detoxifying activity of NZ17074 analogues with multi-layers of selective antimicrobial actions against Escherichia coli and Salmonella enteritidis
Sci Rep, 7, 2017
|
|
5ZO2
 
 | | Crystal structure of mouse nectin-like molecule 4 (mNecl-4) full ectodomain in complex with mouse nectin-like molecule 1 (mNecl-1) Ig1 domain, 3.3A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 3, Cell adhesion molecule 4 | | Authors: | Liu, X, An, T, Li, D, Fan, Z, Xiang, P, Li, C, Ju, W, Li, J, Hu, G, Qin, B, Yin, B, Wojdyla, J.A, Wang, M, Yuan, J, Qiang, B, Shu, P, Cui, S, Peng, X. | | Deposit date: | 2018-04-12 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
4KFW
 
 | |
2LCW
 
 | |
5X9X
 
 | |
2M0Y
 
 | |
6AHZ
 
 | | The NMR Structure of the Polysialyltranseferase Domain (PSTD) in Polysialyltransferase ST8siaIV | | Descriptor: | CMP-N-acetylneuraminate-poly-alpha-2,8-sialyltransferase | | Authors: | Liu, X.H, Lu, B, Peng, L.X, Liao, S.M, Zhou, F, Chen, D, Lu, Z.L, Zhou, G.P, Huang, R.B. | | Deposit date: | 2018-08-21 | | Release date: | 2018-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Inhibition of Polysialyltranseferase ST8SiaIV Through Heparin Binding to Polysialyltransferase Domain (PSTD).
Med Chem, 15, 2019
|
|
6IUX
 
 | | Crystal structure of a hydrolase protein | | Descriptor: | MAGNESIUM ION, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE, [Protein ADP-ribosylarginine] hydrolase | | Authors: | Liu, X.H, Yu, X.C. | | Deposit date: | 2018-12-01 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.195 Å) | | Cite: | Crystal structure of protein
To Be Published
|
|
