6JLE
 
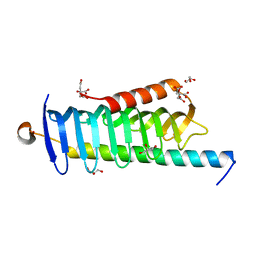 | | Crystal structure of MORN4/Myo3a complex | | Descriptor: | CITRIC ACID, GLYCEROL, MORN repeat-containing protein 4, ... | | Authors: | Li, J, Liu, H, Raval, M.H, Wan, J, Yengo, C.M, Liu, W, Zhang, M. | | Deposit date: | 2019-03-05 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the MORN4/Myo3a Tail Complex Reveals MORN Repeats as Protein Binding Modules.
Structure, 27, 2019
|
|
8U9M
 
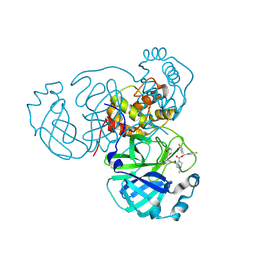 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI95 | | Descriptor: | 3C-like proteinase nsp5, bis(4-fluorophenyl)methyl (1R,2S,5R)-2-({(2R)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}carbamoyl)-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-3-carboxylate | | Authors: | Blankenship, L.R, Liu, W.R. | | Deposit date: | 2023-09-19 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the SARS-CoV-2 main protease in complex with inhibitors
To Be Published
|
|
8U9K
 
 | |
8U9V
 
 | |
8U9T
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI97 | | Descriptor: | (1R,2S,5S)-N~3~,N~3~-bis(4-chlorophenyl)-N~2~-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2,3-dicarboxamide, 3C-like proteinase nsp5 | | Authors: | Blankenship, L.R, Liu, W.R. | | Deposit date: | 2023-09-20 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the SARS-CoV-2 main protease in complex with inhibitors
To Be Published
|
|
8U9N
 
 | |
8U9W
 
 | |
8U9H
 
 | |
8U9U
 
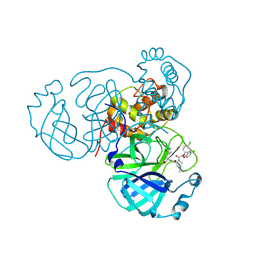 | |
7F3P
 
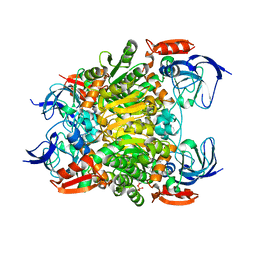 | | Crystal structure of a nadp-dependent alcohol dehydrogenase mutant in apo form | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent isopropanol dehydrogenase, ZINC ION | | Authors: | Han, X, Bi, Y, Wei, H.L, Gao, J, Li, Q, Qu, G, Sun, Z.T, Liu, W.D. | | Deposit date: | 2021-06-16 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Unlocking the Stereoselectivity and Substrate Acceptance of Enzymes: Proline-Induced Loop Engineering Test.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
8WVD
 
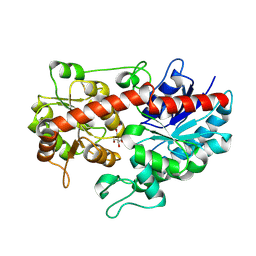 | | Crystal structure of Glycosyltransferase in complex with UD1 | | Descriptor: | Glycosyltransferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Li, J, Shan, N, Yang, J.G, Liu, W.D, Sun, Y.X. | | Deposit date: | 2023-10-23 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Enzymatic Synthesis of Novel Terpenoid Glycoside Derivatives Decorated with N -Acetylglucosamine Catalyzed by UGT74AC1.
J.Agric.Food Chem., 72, 2024
|
|
7WQR
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound 28 | | Descriptor: | Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ~{N}-(3-chlorophenyl)-2-[(3,5-dimethyl-1,2-oxazol-4-yl)methoxy]benzamide | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Sun, H, Fang, P. | | Deposit date: | 2022-01-25 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.124 Å) | | Cite: | Development of highly potent and specific AKR1C3 inhibitors to restore the chemosensitivity of drug-resistant breast cancer.
Eur.J.Med.Chem., 247, 2022
|
|
7WQM
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound 24 | | Descriptor: | 2-[(3,5-dimethyl-1,2-oxazol-4-yl)methoxy]-~{N}-(2-methoxyphenyl)benzamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Sun, H, Fang, P. | | Deposit date: | 2022-01-25 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Development of highly potent and specific AKR1C3 inhibitors to restore the chemosensitivity of drug-resistant breast cancer.
Eur.J.Med.Chem., 247, 2022
|
|
7WQS
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound 25 | | Descriptor: | 2-[(3,5-dimethyl-1,2-oxazol-4-yl)methoxy]-~{N}-(3-methoxyphenyl)benzamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Sun, H, Fang, P. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Development of highly potent and specific AKR1C3 inhibitors to restore the chemosensitivity of drug-resistant breast cancer.
Eur.J.Med.Chem., 247, 2022
|
|
6IRD
 
 | | Complex structure of INADL PDZ89 and PLCb4 C-terminal CC-PBM | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase, GOLD ION, InaD-like protein | | Authors: | Ye, F, Li, J, Huang, Y, Liu, W, Zhang, M. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.813 Å) | | Cite: | An unexpected INAD PDZ tandem-mediated plc beta binding in Drosophila photo receptors.
Elife, 7, 2018
|
|
6IRC
 
 | | C-terminal domain of Drosophila phospholipase b NORPA, methylated | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase | | Authors: | Ye, F, Li, J, Huang, Y, Liu, W, Zhang, M. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-02 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (3.538 Å) | | Cite: | An unexpected INAD PDZ tandem-mediated plc beta binding in Drosophila photo receptors.
Elife, 7, 2018
|
|
5YGK
 
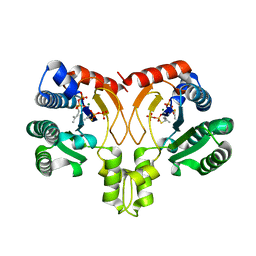 | | Crystal structure of a synthase from Streptomyces sp. CL190 with dmaspp | | Descriptor: | Cyclolavandulyl diphosphate synthase, DIMETHYLALLYL S-THIOLODIPHOSPHATE, MAGNESIUM ION | | Authors: | Gao, J, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-09-23 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Catalytic Role of Conserved Asparagine, Glutamine, Serine, and Tyrosine Residues in Isoprenoid Biosynthesis Enzymes.
Acs Catalysis, 8, 2018
|
|
6LNH
 
 | | Crystal structure of IDO from Bacillus thuringiensis | | Descriptor: | FE (III) ION, L-isoleucine-4-hydroxylase, MERCURY (II) ION | | Authors: | Feng, Y, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-12-30 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of IDO from Bacillus thuringiensis
to be published
|
|
7VPA
 
 | | Crystal structure of Ple629 from marine microbial consortium | | Descriptor: | hydrolase Ple629 | | Authors: | Wu, P, Zhao, Y.P, Li, Z.S, Ingrid, M.C, Lara, P, Gao, J, Han, X, Li, Q, Basak, O, Liu, W.D, Wei, R. | | Deposit date: | 2021-10-15 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular and Biochemical Differences of the Tandem and Cold-Adapted PET Hydrolases Ple628 and Ple629, Isolated From a Marine Microbial Consortium.
Front Bioeng Biotechnol, 10, 2022
|
|
7VMD
 
 | | Crystal structure of a hydrolases Ple628 from marine microbial consortium | | Descriptor: | CALCIUM ION, hydrolase Ple628 | | Authors: | Wu, P, Zhao, Y.P, Li, Z.S, Ingrid, M.C, Lara, P, Gao, J, Han, X, Li, Q, Basak, O, Liu, W.D, Wei, R. | | Deposit date: | 2021-10-08 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular and Biochemical Differences of the Tandem and Cold-Adapted PET Hydrolases Ple628 and Ple629, Isolated From a Marine Microbial Consortium.
Front Bioeng Biotechnol, 10, 2022
|
|
7VPB
 
 | | Crystal structure of a novel hydrolase in apo form | | Descriptor: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, ACETATE ION, plastic degrading hydrolase Ple629 | | Authors: | Wu, P, Zhao, Y.P, Li, Z.S, Ingrid, M.C, Lara, P, Gao, J, Han, X, Li, Q, Basak, O, Liu, W.D, Wei, R. | | Deposit date: | 2021-10-15 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural insight and engineering of a plastic degrading hydrolase Ple629.
Biochem.Biophys.Res.Commun., 626, 2022
|
|
8WAN
 
 | | Structure of transcribing complex 4 (TC4), the initially transcribing complex with Pol II positioned 4nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-07 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (6.07 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
5Z54
 
 | | Crystal structure of a cyclase Hpiu5 from Fischerella sp. ATCC 43239 in complex with cyclo-L-Arg-D-Pro | | Descriptor: | 12-epi-hapalindole C/U synthase, CALCIUM ION, amino({3-[(3S,8aS)-1,4-dioxooctahydropyrrolo[1,2-a]pyrazin-3-yl]propyl}amino)methaniminium | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-01-17 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Z53
 
 | | Crystal structure of a cyclase Filc from Fischerella sp. in complex with cyclo-L-Arg-D-Pro | | Descriptor: | 12-epi-hapalindole U synthase, CALCIUM ION, SULFATE ION, ... | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-01-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6KA2
 
 | | Crystal structure of a Thebaine synthase from Papaver somniferum in complex with TBN | | Descriptor: | (4R,7aR,12bS)-7,9-dimethoxy-3-methyl-2,4,7a,13-tetrahydro-1H-4,12-methanobenzofuro[3,2-e]isoquinoline, Thebaine synthase 2 | | Authors: | Xue, J, Yu, X.J, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-06-20 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into thebaine synthase 2 catalysis.
Biochem.Biophys.Res.Commun., 529, 2020
|
|
