4RML
 
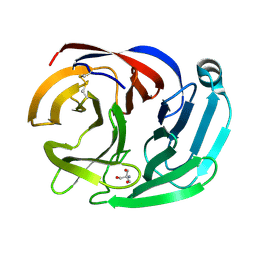 | | Crystal structure of the Olfactomedin domain of latrophilin 3 in C2221 crystal form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Latrophilin-3, MAGNESIUM ION | | Authors: | Ranaivoson, F.M, Liu, Q, Martini, F, Bergami, F, Von daake, S, Li, S, Demeler, B, Hendrickson, W.A, Comoletti, D. | | Deposit date: | 2014-10-21 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and Mechanistic Insights into the Latrophilin3-FLRT3 Complex that Mediates Glutamatergic Synapse Development.
Structure, 23, 2015
|
|
3QB5
 
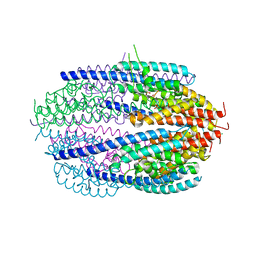 | | Human C3PO complex in the presence of MnSO4 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Ye, X, Huang, N, Liu, Y, Paroo, Z, Chen, S, Zhang, H, Liu, Q. | | Deposit date: | 2011-01-12 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of C3PO and mechanism of human RISC activation.
Nat.Struct.Mol.Biol., 18, 2011
|
|
8HB9
 
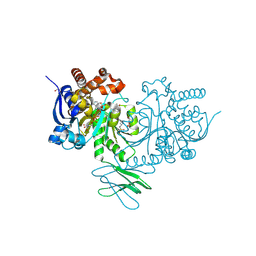 | | Crystal Structure of Human IDH1 R132H Mutant in Complex with NADPH and Compound IHMT-IDH1-053 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Guo, G, Wang, B, Liu, J, Liu, Q. | | Deposit date: | 2022-10-27 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based discovery of IHMT-IDH1-053 as a potent irreversible IDH1 mutant selective inhibitor.
Eur.J.Med.Chem., 256, 2023
|
|
4IQ8
 
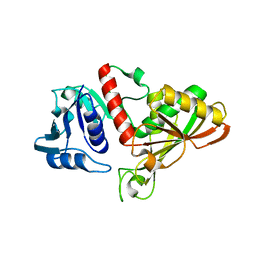 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 3 from Saccharomyces cerevisiae | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase 3 | | Authors: | Wang, H, Liu, Q, Niu, L, Teng, M, Li, X. | | Deposit date: | 2013-01-11 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Preliminary crystallographic analysis of glyceraldehyde-3-phosphate dehydrogenase 3 from Saccharomyces cerevisiae.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
1Y00
 
 | | Solution structure of the Carbon Storage Regulator protein CsrA | | Descriptor: | Carbon storage regulator | | Authors: | Gutierrez, P, Li, Y, Osborne, M.J, Liu, Q, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2004-11-13 | | Release date: | 2005-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the carbon storage regulator protein CsrA from Escherichia coli.
J.Bacteriol., 187, 2005
|
|
3QIO
 
 | | Crystal Structure of HIV-1 RNase H with engineered E. coli loop and N-hydroxy quinazolinedione inhibitor | | Descriptor: | 3-hydroxy-6-(phenylsulfonyl)quinazoline-2,4(1H,3H)-dione, Gag-Pol polyprotein,Ribonuclease HI,Gag-Pol polyprotein, MANGANESE (II) ION, ... | | Authors: | Lansdon, E.B, Liu, Q. | | Deposit date: | 2011-01-27 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4011 Å) | | Cite: | Structural and Binding Analysis of Pyrimidinol Carboxylic Acid and N-Hydroxy Quinazolinedione HIV-1 RNase H Inhibitors.
Antimicrob.Agents Chemother., 55, 2011
|
|
2QXL
 
 | | Crystal Structure Analysis of Sse1, a yeast Hsp110 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein homolog SSE1, MAGNESIUM ION, ... | | Authors: | Hendrickson, W.A, Liu, Q. | | Deposit date: | 2007-08-12 | | Release date: | 2007-10-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Insights into hsp70 chaperone activity from a crystal structure of the yeast hsp110 Sse1.
Cell(Cambridge,Mass.), 131, 2007
|
|
8F6T
 
 | | Cryo-EM structure of alkane 1-monooxygenase AlkB-AlkG complex from Fontimonas thermophila | | Descriptor: | Alkane 1-monooxygenase, DODECANE, FE (III) ION | | Authors: | Chai, J, Guo, G, McSweeney, S, Shanklin, J, Liu, Q. | | Deposit date: | 2022-11-17 | | Release date: | 2023-04-05 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis for enzymatic terminal C-H bond functionalization of alkanes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2INF
 
 | | Crystal Structure of Uroporphyrinogen Decarboxylase from Bacillus subtilis | | Descriptor: | Uroporphyrinogen decarboxylase | | Authors: | Fan, J, Liu, Q, Hao, Q, Teng, M.K, Niu, L.W. | | Deposit date: | 2006-10-06 | | Release date: | 2006-10-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of uroporphyrinogen decarboxylase from Bacillus subtilis
J.Bacteriol., 189, 2007
|
|
7SEV
 
 | | Crystal structure of E coli contaminant protein YadF co-purified with a plant protein | | Descriptor: | Carbonic anhydrase 2, POTASSIUM ION, ZINC ION | | Authors: | Chai, L, Zhu, P, Chai, J, Pang, C, Andi, B, McsWeeney, S, Shanklin, J, Liu, Q. | | Deposit date: | 2021-10-01 | | Release date: | 2021-11-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | AlphaFold Protein Structure Database for Sequence-Independent Molecular Replacement
Crystals, 11, 2021
|
|
7SEU
 
 | | Crystal structure of E coli contaminant protein YncE co-purified with a plant protein | | Descriptor: | PQQ-dependent catabolism-associated beta-propeller protein | | Authors: | Chai, L, Zhu, P, Chai, J, Pang, C, Andi, B, McsWeeney, S, Shanklin, J, Liu, Q. | | Deposit date: | 2021-10-01 | | Release date: | 2022-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | AlphaFold Protein Structure Database for Sequence-Independent Molecular Replacement
Crystals, 11, 2021
|
|
4YEB
 
 | | Structural characterization of a synaptic adhesion complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Fibronectin leucine rich transmembrane protein 3, ... | | Authors: | Ranaivoson, F.M, Liu, Q, Martini, F, Bergami, F, von Daake, S, Li, S, Lee, D, Demeler, B, Hendrickson, W.A, Comoletti, D. | | Deposit date: | 2015-02-23 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structural and Mechanistic Insights into the Latrophilin3-FLRT3 Complex that Mediates Glutamatergic Synapse Development.
Structure, 23, 2015
|
|
6O8A
 
 | | Thaumatin native-SAD structure determined at 5 keV from microcrystals | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Guo, G, Zhu, P, Fuchs, M.R, Shi, W, Andi, B, Gao, Y, Hendrickson, W.A, McSweeney, S, Liu, Q. | | Deposit date: | 2019-03-09 | | Release date: | 2019-05-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synchrotron microcrystal native-SAD phasing at a low energy.
Iucrj, 6, 2019
|
|
4MMJ
 
 | | crystal structure of YafQ from E.coli strain BL21(DE3) | | Descriptor: | Addiction module toxin, RelE/StbE family, SULFATE ION | | Authors: | Liang, Y, Gao, Z, Liu, Q, Dong, Y. | | Deposit date: | 2013-09-09 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Characterization of Escherichia coli Toxin-Antitoxin Complex DinJ-YafQ
J.Biol.Chem., 289, 2014
|
|
5GNI
 
 | |
6IUP
 
 | | Crystal structure of FGFR4 kinase domain in complex with compound 5 | | Descriptor: | DIMETHYL SULFOXIDE, Fibroblast growth factor receptor 4, N-{4-[4-amino-3-(3,5-dimethyl-1-benzofuran-2-yl)-7-oxo-6,7-dihydro-2H-pyrazolo[3,4-d]pyridazin-2-yl]phenyl}prop-2-enamide | | Authors: | Xu, Y, Liu, Q. | | Deposit date: | 2018-11-29 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
4PQZ
 
 | |
6D9Z
 
 | | Structure of CysZ, a sulfate permease from Pseudomonas Denitrificans | | Descriptor: | Sulfate transporter CysZ, octyl beta-D-glucopyranoside | | Authors: | Sanghai, Z.A, Clarke, O.B, Liu, Q, Banerjee, S, Rajashankar, K.R, Hendrickson, W.A, Mancia, F. | | Deposit date: | 2018-04-30 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.4021318 Å) | | Cite: | Structure-based analysis of CysZ-mediated cellular uptake of sulfate.
Elife, 7, 2018
|
|
6IUO
 
 | | Crystal structure of FGFR4 kinase domain in complex with a covalent inhibitor | | Descriptor: | Fibroblast growth factor receptor 4, N-({4-[4-amino-3-(3,5-dimethyl-1-benzofuran-2-yl)-7-oxo-6,7-dihydro-2H-pyrazolo[3,4-d]pyridazin-2-yl]phenyl}methyl)prop-2-enamide | | Authors: | Xu, Y, Liu, Q. | | Deposit date: | 2018-11-29 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
6ITJ
 
 | | Crystal structure of FGFR1 kinase domain in complex with compound 3 | | Descriptor: | 4-azanyl-3-(3,5-dimethyl-1-benzofuran-2-yl)-2-phenyl-6~{H}-pyrazolo[3,4-d]pyridazin-7-one, Fibroblast growth factor receptor 1 | | Authors: | Xu, Y, Liu, Q. | | Deposit date: | 2018-11-23 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
3DXX
 
 | | Crystal structure of EcTrmB | | Descriptor: | GLYCEROL, tRNA (guanine-N(7)-)-methyltransferase | | Authors: | Zhou, H.H, Liu, Q, Gao, Y.X, Teng, M.K, Niu, L.W. | | Deposit date: | 2008-07-25 | | Release date: | 2009-05-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Monomeric tRNA (m(7)G46) methyltransferase from Escherichia coli presents a novel structure at the function-essential insertion
Proteins, 76, 2009
|
|
3EWG
 
 | | Crystal structure of the N-terminal domain of NusG (NGN) from Methanocaldococcus jannaschii | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Putative transcription antitermination protein nusG | | Authors: | Zhou, H.H, Liu, Q, Gao, Y.X, Teng, M.K, Niu, L.W. | | Deposit date: | 2008-10-15 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of NusG N-terminal (NGN) domain from Methanocaldococcus jannaschii and its interaction with rpoE''
Proteins, 76, 2009
|
|
3DXZ
 
 | | Crystal structure of EcTrmB in complex with SAH | | Descriptor: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Zhou, H.H, Liu, Q, Gao, Y.X, Teng, M.K, Niu, L.W. | | Deposit date: | 2008-07-25 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Monomeric tRNA (m(7)G46) methyltransferase from Escherichia coli presents a novel structure at the function-essential insertion
Proteins, 76, 2009
|
|
3DXY
 
 | | Crystal structure of EcTrmB in complex with SAM | | Descriptor: | GLYCEROL, S-ADENOSYLMETHIONINE, SULFATE ION, ... | | Authors: | Zhou, H.H, Liu, Q, Gao, Y.X, Teng, M.K, Niu, L.W. | | Deposit date: | 2008-07-25 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Monomeric tRNA (m(7)G46) methyltransferase from Escherichia coli presents a novel structure at the function-essential insertion
Proteins, 76, 2009
|
|
8ENA
 
 | | Thaumatin native-SAD structure determined at 5 keV with a helium environmet | | Descriptor: | Thaumatin-1 | | Authors: | Karasawa, A, Andi, B, Ruchs, M.R, Shi, W, McSweeney, S, Hendrickson, W.A, Liu, Q. | | Deposit date: | 2022-09-29 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multi-crystal native-SAD phasing at 5 keV with a helium environment.
Iucrj, 9, 2022
|
|
