8WQR
 
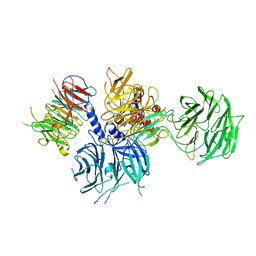 | | Structure of the DDB1-AMBRA1 E3 ligase receptor complex linked to cell cycle regulation | | Descriptor: | Activating molecule in BECN1-regulated autophagy protein 1, DNA damage-binding protein 1 | | Authors: | Liu, M, Wang, Y, Su, M.Y, Stjepanovic, G. | | Deposit date: | 2023-10-12 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structure of the DDB1-AMBRA1 E3 ligase receptor complex linked to cell cycle regulation.
Nat Commun, 14, 2023
|
|
6KXG
 
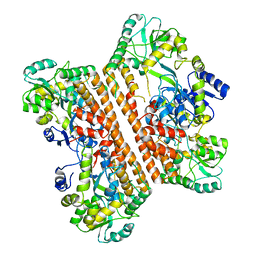 | | Crystal structure of caspase-11-CARD | | Descriptor: | caspase-11-CARD | | Authors: | Liu, M.Z.Y, Jin, T.C. | | Deposit date: | 2019-09-11 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Crystal structure of caspase-11 CARD provides insights into caspase-11 activation.
Cell Discov, 6, 2020
|
|
6KY4
 
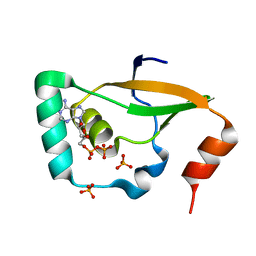 | | Crystal structure of Sulfiredoxin from Arabidopsis thaliana | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, Sulfiredoxin, ... | | Authors: | Liu, M, Wang, J, Li, X, Li, M, Sylvanno, M.J, Zhang, M, Wang, M. | | Deposit date: | 2019-09-16 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of sulfiredoxin from Arabidopsis thaliana revealed a more robust antioxidant mechanism in plants.
Biochem.Biophys.Res.Commun., 520, 2019
|
|
8GVD
 
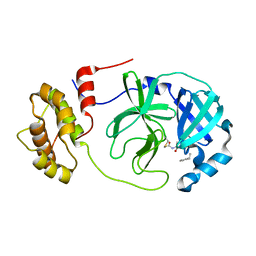 | | SARS-CoV-2 Mpro in complex with D-4-38 | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, 3C-like proteinase nsp5 | | Authors: | Liu, M, Huang, H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SARS-CoV-2 Mpro in complex with D-4-38
To Be Published
|
|
8GVY
 
 | | SARS CoV-2 Mpro in complex with D-3-149 | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, 3C-like proteinase nsp5 | | Authors: | Liu, M, Huang, H. | | Deposit date: | 2022-09-16 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | SARS CoV-2 Mpro in complex with D-3-149
To Be Published
|
|
8GWJ
 
 | |
7C39
 
 | | Crystal structure of AofleA from Arthrobotrys oligospora in complex with methylated L-fucose | | Descriptor: | AoflcA, CITRIC ACID, GLYCEROL, ... | | Authors: | Liu, M, Cheng, X, Wang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-05-11 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insights into the fungi-nematodes interaction mediated by fucose-specific lectin AofleA from Arthrobotrys oligospora.
Int.J.Biol.Macromol., 164, 2020
|
|
7C3D
 
 | | Crystal structure of AofleA from Arthrobotrys oligospora in complex with D-arabinose | | Descriptor: | AofleA, GLYCEROL, alpha-D-arabinopyranose, ... | | Authors: | Liu, M, Cheng, X, Wang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-05-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.596 Å) | | Cite: | Structural insights into the fungi-nematodes interaction mediated by fucose-specific lectin AofleA from Arthrobotrys oligospora.
Int.J.Biol.Macromol., 164, 2020
|
|
7C38
 
 | | Crystal structure of AofleA from Arthrobotrys oligospora in complex with L-fucose | | Descriptor: | AofleA, GLYCEROL, alpha-L-fucopyranose, ... | | Authors: | Liu, M, Cheng, X, Wang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-05-11 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural insights into the fungi-nematodes interaction mediated by fucose-specific lectin AofleA from Arthrobotrys oligospora.
Int.J.Biol.Macromol., 164, 2020
|
|
7C3E
 
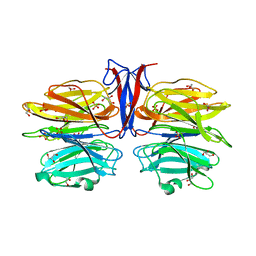 | | Crystal structure of R97A/R150A/R203A mutant of AofleA from Arthrobotrys oligospora | | Descriptor: | AofleA, GLYCEROL, SULFATE ION | | Authors: | Liu, M, Cheng, X, Wang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-05-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.183 Å) | | Cite: | Structural insights into the fungi-nematodes interaction mediated by fucose-specific lectin AofleA from Arthrobotrys oligospora.
Int.J.Biol.Macromol., 164, 2020
|
|
7C3C
 
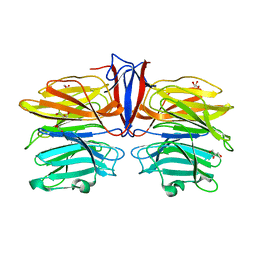 | | Crystal structure of AofleA from Arthrobotrys oligospora in complex with D-manose | | Descriptor: | AofleA, GLYCEROL, alpha-D-mannopyranose, ... | | Authors: | Liu, M, Cheng, X, Wang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-05-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.301 Å) | | Cite: | Structural insights into the fungi-nematodes interaction mediated by fucose-specific lectin AofleA from Arthrobotrys oligospora.
Int.J.Biol.Macromol., 164, 2020
|
|
7C37
 
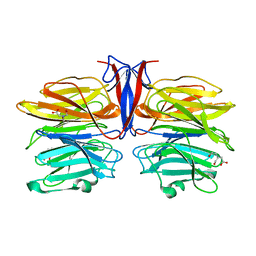 | | Crystal structure of AofleA from Arthrobotrys oligospora | | Descriptor: | AofleA, BICINE | | Authors: | Liu, M, Cheng, X, Wang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-05-11 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Structural insights into the fungi-nematodes interaction mediated by fucose-specific lectin AofleA from Arthrobotrys oligospora.
Int.J.Biol.Macromol., 164, 2020
|
|
8CQP
 
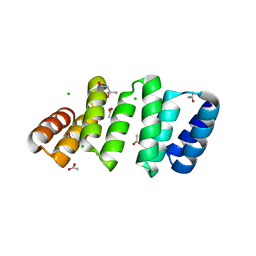 | | Crystal structure of an 8-repeat consensus TPR superhelix with Calcium (low concentration) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Liutkus, M, Rojas, A.L, Cortajarena, A.L. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Diverse crystalline protein scaffolds through metal-dependent polymorphism.
Protein Sci., 33, 2024
|
|
8CQQ
 
 | |
7Q34
 
 | | Crystal structure of the multidrug binding transcriptional regulator LmrR in complex squaraine dye | | Descriptor: | 2,4-bis[(E)-(1-ethyl-3,3-dimethyl-indol-2-ylidene)methyl]cyclobutane-1,3-dione, Helix-turn-helix transcriptional regulator, NICKEL (II) ION | | Authors: | Liutkus, M, Mejias, S.H, Barolo, C, Cortajarena, A.L. | | Deposit date: | 2021-10-26 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Designing Artificial Fluorescent Proteins: Squaraine-LmrR Biophosphors for High Performance Deep-Red Biohybrid Light-Emitting Diodes
Adv Funct Mater, 32, 2022
|
|
8OT7
 
 | |
8BU0
 
 | | Crystal structure of an 8 repeat consensus TPR superhelix with calcium | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Liutkus, M, Rojas, A.L, Cortajarena, A.L. | | Deposit date: | 2022-11-30 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diverse crystalline protein scaffolds through metal-dependent polymorphism.
Protein Sci., 33, 2024
|
|
8CHY
 
 | | Crystal structure of an 8-repeat consensus TPR superhelix with Zinc. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Liutkus, M, Rojas, A.L, Cortajarena, A.L. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diverse crystalline protein scaffolds through metal-dependent polymorphism.
Protein Sci., 33, 2024
|
|
8CIG
 
 | | Crystal structure of an 8-repeat consensus TPR superhelix in tris Buffer with Calcium. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Liutkus, M, Rojas, A.L, Cortajarena, A.L. | | Deposit date: | 2023-02-09 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diverse crystalline protein scaffolds through metal-dependent polymorphism.
Protein Sci., 33, 2024
|
|
8CP8
 
 | | Crystal structure of an 8-repeat consensus TPR superhelix with Lead | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Liutkus, M, Rojas, A.L, Cortajarena, A.L. | | Deposit date: | 2023-03-02 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Diverse crystalline protein scaffolds through metal-dependent polymorphism.
Protein Sci., 33, 2024
|
|
8CH0
 
 | | Crystal structure of an 8-repeat consensus TPR superhelix with Gadolinium. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Consensus tetratricopeptide repeat protein, ... | | Authors: | Liutkus, M, Rojas, A.L, Cortajarena, A.L. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Diverse crystalline protein scaffolds through metal-dependent polymorphism.
Protein Sci., 33, 2024
|
|
8CKR
 
 | | Crystal structure of an 8-repeat consensus TPR superhelix with in Hepes with Ca | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Consensus tetratricopeptide repeat protein | | Authors: | Liutkus, M, Rojas, A.L, Cortajarena, A.L. | | Deposit date: | 2023-02-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Diverse crystalline protein scaffolds through metal-dependent polymorphism.
Protein Sci., 33, 2024
|
|
8CMQ
 
 | | Crystal structure of an 8-repeat consensus TPR superhelix with Tb | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Consensus tetratricopeptide repeat protein, ... | | Authors: | Liutkus, M, Rojas, A.L, Cortajarena, A.L. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.782 Å) | | Cite: | Diverse crystalline protein scaffolds through metal-dependent polymorphism.
Protein Sci., 33, 2024
|
|
3JSD
 
 | | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Weiss, M.A, Wan, Z.L, Dodson, E.J, Liu, M, Xu, B, Hua, Q.X, Turkenburg, M, Whittingham, J, Nakagawa, S.H, Huang, K, Hu, S.Q, Jia, W.H, Wang, S.H, Brange, J, Whittaker, J, Arvan, P, Katsoyannis, P.G, Dodson, G.G. | | Deposit date: | 2009-09-10 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus
To be Published
|
|
3BH4
 
 | | High resolution crystal structure of Bacillus amyloliquefaciens alpha-amylase | | Descriptor: | Alpha-amylase, CALCIUM ION, SODIUM ION | | Authors: | Alikhajeh, J, Khajeh, K, Ranjbar, B, Naderi-Manesh, H, Lin, Y.H, Liu, M.Y, Chen, C.J. | | Deposit date: | 2007-11-28 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of Bacillus amyloliquefaciens alpha-amylase at high resolution: implications for thermal stability.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
