6UAD
 
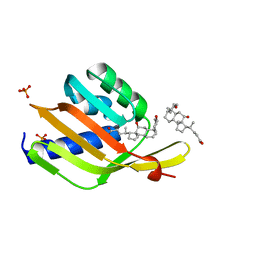 | | Ketosteroid isomerase (C. testosteroni) with truncated & designed loop for precise positioning of a catalytic E38 | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, Ketosteroid isomerase with truncated and designed loop, PHOSPHATE ION | | Authors: | Kundert, K, Thompson, M.C, Liu, L, Fraser, J.S, Kortemme, T. | | Deposit date: | 2019-09-10 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Ketosteroid isomerase (C. testosteroni) with truncated & designed loop for precise positioning of a catalytic E38
To Be Published
|
|
8E61
 
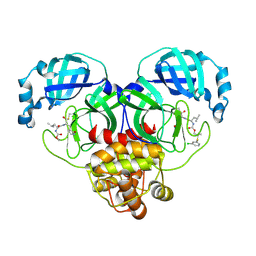 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a m-chlorophenyl dimethyl sulfane inhibitor | | Descriptor: | (1R,2S)-2-{[N-({2-[(3-chlorophenyl)sulfanyl]-2-methylpropoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({2-[(3-chlorophenyl)sulfanyl]-2-methylpropoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Madden, T.K, Groutas, W.C. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
8E69
 
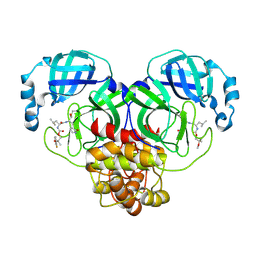 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a m-fluorodimethyl oxybenzene inhibitor | | Descriptor: | (1R,2S)-2-[(N-{[2-(3-fluorophenoxy)-2-methylpropoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Miller, M.J, Groutas, W.C. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
8E6A
 
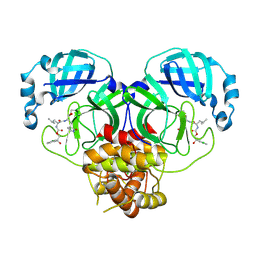 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a p-chlorophenylethanol based inhibitor | | Descriptor: | (1R,2S)-2-[(N-{[(2S)-2-(3-chlorophenyl)-2-hydroxypropoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl]propane-1-sulfonic acid, (1S,2S)-2-[(N-{[(2R)-2-(3-chlorophenyl)-2-hydroxypropoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
8E64
 
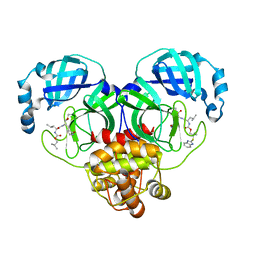 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a benzimidazole dimethyl sulfane inhibitor | | Descriptor: | (1S,2S)-2-{[N-({2-[(1H-benzimidazol-2-yl)sulfanyl]-2-methylpropoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1~{R},2~{S})-2-[[(2~{S})-2-[[2-(1~{H}-benzimidazol-2-ylsulfanyl)-2-methyl-propoxy]carbonylamino]-4-methyl-pentanoyl]amino]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propane-1-sulfonic acid;molecular oxygen, 3C-like proteinase | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Madden, T.K, Groutas, W.C. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
8E5Z
 
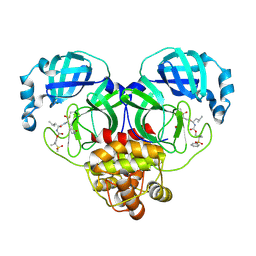 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a dimethyl sulfonyl benzene inhibitor | | Descriptor: | (1R,2S)-2-[(N-{[2-(benzenesulfonyl)-2-methylpropoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-[(N-{[2-(benzenesulfonyl)-2-methylpropoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Miller, M.J, Groutas, W.C. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
8E65
 
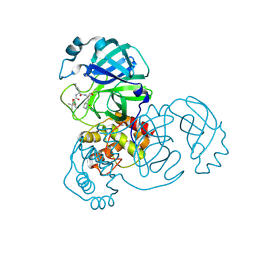 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a p-chlorodimethyl oxybenzene inhibitor | | Descriptor: | (1S,2S)-2-[(N-{[2-(4-chlorophenoxy)-2-methylpropoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Miller, M.J, Groutas, W.C. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
8E68
 
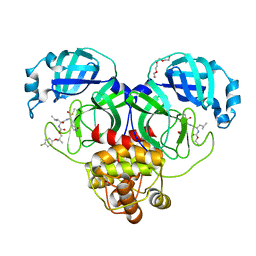 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a p-fluorodimethyl oxybenzene inhibitor | | Descriptor: | (1S,2S)-2-[(N-{[2-(4-fluorophenoxy)-2-methylpropoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, N~2~-{[2-(4-fluorophenoxy)-2-methylpropoxy]carbonyl}-N-{(1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-1-sulfanylpropan-2-yl}-L-leucinamide, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Miller, M.J, Groutas, W.C. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
8E63
 
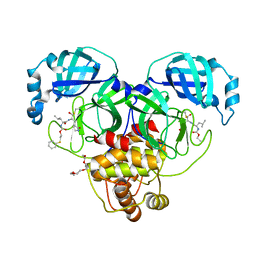 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a phenyl sulfane inhibitor | | Descriptor: | (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-[(N-{[2-(phenylsulfanyl)ethoxy]carbonyl}-L-leucyl)amino]propane-1-sulfonic acid, 2-phenylsulfanylethyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Madden, T.K, Groutas, W.C. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
4R8X
 
 | | Crystal structure of a uricase from Bacillus fastidious | | Descriptor: | Uricase | | Authors: | Feng, J, Wang, L, Liu, H.B, Liu, L, Liao, F. | | Deposit date: | 2014-09-03 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Crystal structure of Bacillus fastidious uricase reveals an unexpected folding of the C-terminus residues crucial for thermostability under physiological conditions.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
6P5S
 
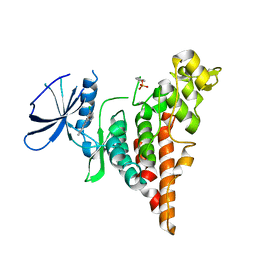 | | HIPK2 kinase domain bound to CX-4945 | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Homeodomain-interacting protein kinase 2 | | Authors: | Agnew, C, Liu, L, Jura, N. | | Deposit date: | 2019-05-30 | | Release date: | 2019-07-31 | | Last modified: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | The crystal structure of the protein kinase HIPK2 reveals a unique architecture of its CMGC-insert region.
J.Biol.Chem., 294, 2019
|
|
8IQ6
 
 | | Cryo-EM structure of Latanoprost-bound prostaglandin-F2-alpha receptor-miniGq-Nb35 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Lv, X, Gao, K, Nie, J, Zhang, X, Zhang, S, Ren, Y, Li, Q, Huang, J, Liu, L, Zhang, X, Sun, X, Zhang, W, Liu, X. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of human prostaglandin F 2 alpha receptor reveal the mechanism of ligand and G protein selectivity.
Nat Commun, 14, 2023
|
|
8IQ4
 
 | | Cryo-EM structure of Carboprost-bound prostaglandin-F2-alpha receptor-miniGq-Nb35 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Lv, X, Gao, K, Nie, J, Zhang, X, Zhang, S, Ren, Y, Li, Q, Huang, J, Liu, L, Zhang, X, Sun, X, Zhang, W, Liu, X. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of human prostaglandin F 2 alpha receptor reveal the mechanism of ligand and G protein selectivity.
Nat Commun, 14, 2023
|
|
8AZU
 
 | | Paired helical tau filaments from high-spin supernatants of aqueous extracts from Alzheimer's disease brains | PHF Tau | | Descriptor: | Microtubule-associated protein tau | | Authors: | Yang, Y, Stern, M.A, Meunier, L.A, Liu, W, Cai, Y.Q, Ericsson, M, Liu, L, Selkoe, J.D, Goedert, M, Scheres, H.W.S. | | Deposit date: | 2022-09-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Abundant A beta fibrils in ultracentrifugal supernatants of aqueous extracts from Alzheimer's disease brains.
Neuron, 111, 2023
|
|
5WLK
 
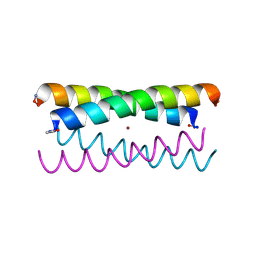 | |
5WZE
 
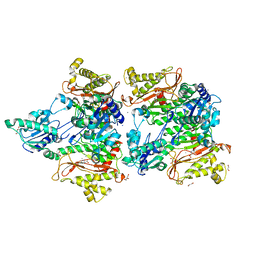 | | The structure of Pseudomonas aeruginosa aminopeptidase PepP | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, Aminopeptidase P, ... | | Authors: | Bao, R, Peng, C.T, Liu, L, He, L.H, Li, C.C, Li, T, Shen, Y.L, Zhu, Y.B, Song, Y.J. | | Deposit date: | 2017-01-17 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Structure-Function Relationship of Aminopeptidase P from Pseudomonas aeruginosa.
Front Microbiol, 8, 2017
|
|
9BTS
 
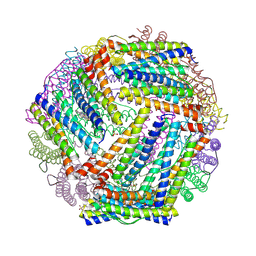 | | Crystal structure of the bacterioferritin (Bfr) and ferritin (Ftn) heterooligomer complex from Acinetobacter baumannii | | Descriptor: | Bacterioferritin (Bfr), Ferritin (Ftn), PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Rivera, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of Acinetobacter baumannii bacterioferritin reveals a heteropolymer of bacterioferritin and ferritin subunits.
Sci Rep, 14, 2024
|
|
7BOR
 
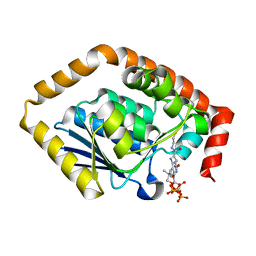 | | Structure of Pseudomonas aeruginosa CoA-bound OdaA | | Descriptor: | COENZYME A, Probable enoyl-CoA hydratase/isomerase | | Authors: | Zhao, N, Zhao, C, Liu, L, Li, T, Li, C, He, L, Zhu, Y, Song, Y, Bao, R. | | Deposit date: | 2020-03-19 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural and molecular dynamic studies of Pseudomonas aeruginosa OdaA reveal the regulation role of a C-terminal hinge element.
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
9IPU
 
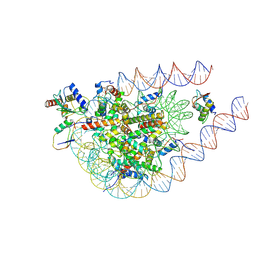 | |
8E7B
 
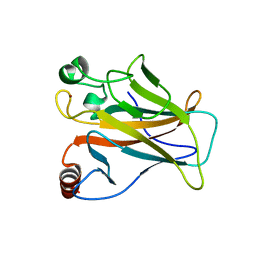 | | Crystal structure of the p53 (Y107H) core domain monoclinic P form | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Miller, S, Karanicolas, J. | | Deposit date: | 2022-08-23 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An African-Specific Variant of TP53 Reveals PADI4 as a Regulator of p53-Mediated Tumor Suppression.
Cancer Discov, 13, 2023
|
|
8E7A
 
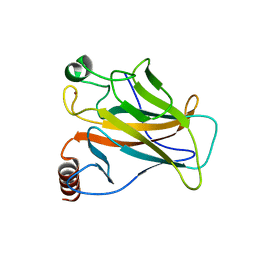 | | Crystal structure of the p53 (Y107H) core domain orthorhombic P form | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Miller, S, Karanicolas, J. | | Deposit date: | 2022-08-23 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An African-Specific Variant of TP53 Reveals PADI4 as a Regulator of p53-Mediated Tumor Suppression.
Cancer Discov, 13, 2023
|
|
8IS0
 
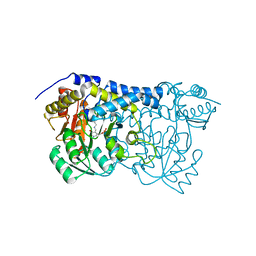 | | Carbon Sulfoxide lyase - Y106F | | Descriptor: | 2-AMINO-ACRYLIC ACID, PYRIDOXAL-5'-PHOSPHATE, Probable hercynylcysteine sulfoxide lyase | | Authors: | Gong, W.M, Wei, L.L, Liu, L. | | Deposit date: | 2023-03-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structure of mycobacterial ergothioneine-biosynthesis C-S lyase EgtE.
J.Biol.Chem., 300, 2024
|
|
8IRK
 
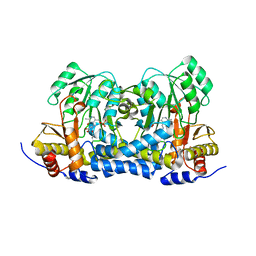 | | Carbon Sulfoxide lyase | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, PYRUVIC ACID, Probable hercynylcysteine sulfoxide lyase | | Authors: | Gong, W.M, Wei, L.L, Liu, L. | | Deposit date: | 2023-03-18 | | Release date: | 2024-03-06 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of mycobacterial ergothioneine-biosynthesis C-S lyase EgtE.
J.Biol.Chem., 300, 2024
|
|
8IRY
 
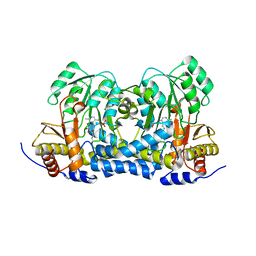 | | Carbon Sulfoxide lyase | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, PYRUVIC ACID, Probable hercynylcysteine sulfoxide lyase | | Authors: | Gong, W.M, Wei, L.L, Liu, L. | | Deposit date: | 2023-03-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of mycobacterial ergothioneine-biosynthesis C-S lyase EgtE.
J.Biol.Chem., 300, 2024
|
|
8IRZ
 
 | | Carbon Sulfoxide lyase | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Probable hercynylcysteine sulfoxide lyase | | Authors: | Gong, W.M, Wei, L.L, Liu, L. | | Deposit date: | 2023-03-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structure of mycobacterial ergothioneine-biosynthesis C-S lyase EgtE.
J.Biol.Chem., 300, 2024
|
|
