4YLC
 
 | |
4YLB
 
 | |
8XR5
 
 | | Crystal structure of PD-L1 complexed with small molecule inhibitor X18 | | Descriptor: | (2~{R})-2-[[2-(2,1,3-benzoxadiazol-5-ylmethoxy)-5-chloranyl-4-[[2-fluoranyl-3-[3-[3-(4-oxidanylpiperidin-1-yl)propoxy]phenyl]phenyl]methoxy]phenyl]methylamino]-3-oxidanyl-propanoic acid, Programmed cell death 1 ligand 1 | | Authors: | Liu, L. | | Deposit date: | 2024-01-06 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of Novel PD-L1 Small-Molecular Inhibitors with Potent In Vivo Anti-tumor Immune Activity.
J.Med.Chem., 67, 2024
|
|
1IU9
 
 | | Crystal structure of the C-terminal domain of aspartate racemase from Pyrococcus horikoshii OT3 | | Descriptor: | CALCIUM ION, aspartate racemase | | Authors: | Liu, L, Iwata, K, Yohda, M, Miki, K. | | Deposit date: | 2002-02-28 | | Release date: | 2003-09-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural insight into gene duplication, gene fusion and domain swapping in the evolution of PLP-independent amino acid racemases
FEBS LETT., 528, 2002
|
|
6M4J
 
 | | SspA in complex with cysteine | | Descriptor: | CYSTEINE, PYRIDOXAL-5'-PHOSPHATE, SspA complex protein | | Authors: | Liu, L, Gao, H. | | Deposit date: | 2020-03-07 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of an l-Cysteine Desulfurase from an Ssp DNA Phosphorothioation System.
Mbio, 11, 2020
|
|
1IUA
 
 | | Ultra-high resolution structure of HiPIP from Thermochromatium tepidum | | Descriptor: | High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Liu, L, Nogi, T, Kobayashi, M, Nozawa, T, Miki, K. | | Deposit date: | 2002-03-01 | | Release date: | 2002-03-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Ultrahigh-resolution structure of high-potential iron-sulfur protein from Thermochromatium tepidum.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3HH3
 
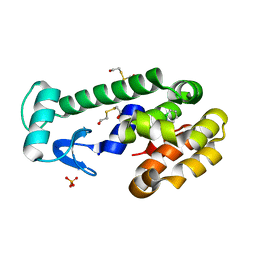 | | New azaborine compounds bind to the T4 lysozyme L99A cavity - 1,2-dihydro-1,2-azaborine | | Descriptor: | 1,2-dihydro-1,2-azaborinine, 2-HYDROXYETHYL DISULFIDE, Lysozyme, ... | | Authors: | Liu, L, Matthews, B.W. | | Deposit date: | 2009-05-14 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Boron mimetics: 1,2-dihydro-1,2-azaborines bind inside a nonpolar cavity of T4 lysozyme.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3HH4
 
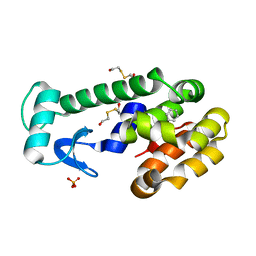 | | New azaborine compounds bind to the T4 lysozyme L99A cavity - Benzene as control | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, BENZENE, Lysozyme, ... | | Authors: | Liu, L, Matthews, B.W. | | Deposit date: | 2009-05-14 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Boron mimetics: 1,2-dihydro-1,2-azaborines bind inside a nonpolar cavity of T4 lysozyme.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3HH5
 
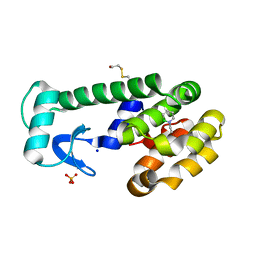 | | New azaborine compounds bind to the T4 lysozyme L99A cavity - 1-ethyl-2-hydro-1,2-azaborine | | Descriptor: | 1-ethyl-1,2-dihydro-1,2-azaborinine, 2-HYDROXYETHYL DISULFIDE, Lysozyme, ... | | Authors: | Liu, L, Matthews, B.W. | | Deposit date: | 2009-05-14 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Boron mimetics: 1,2-dihydro-1,2-azaborines bind inside a nonpolar cavity of T4 lysozyme.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3HH6
 
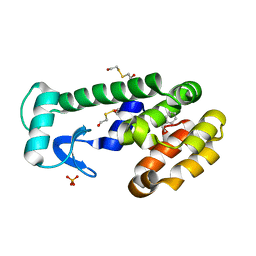 | | New azaborine compounds bind to the T4 lysozyme L99A cavity -ethylbenzene as control | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, Lysozyme, PHENYLETHANE, ... | | Authors: | Liu, L, Matthews, B.W. | | Deposit date: | 2009-05-14 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Boron mimetics: 1,2-dihydro-1,2-azaborines bind inside a nonpolar cavity of T4 lysozyme.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
8CZV
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 17d | | Descriptor: | 3C-like proteinase, [(1~{S},2~{R})-2-[4,4-bis(fluoranyl)cyclohexyl]cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, [(1~{S},2~{R})-2-[4,4-bis(fluoranyl)cyclohexyl]cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8CZU
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 16d | | Descriptor: | 3C-like proteinase, [(1~{S},2~{S})-2-(cyclohexylmethyl)cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8CZT
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 15d | | Descriptor: | 3C-like proteinase, [(1~{S},2~{R})-2-cyclohexylcyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
6C4X
 
 | | Cross-alpha Amyloid-like Structure alphaAmmem | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ZINC ION, cross-alpha amyloid-like membrane peptide alpha-AmMEM | | Authors: | Liu, L, Zhang, S.Q. | | Deposit date: | 2018-01-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
6C50
 
 | | Cross-alpha Amyloid-like Structure alphaAmS | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cross-alpha Amyloid-like Structure alphaAmS, FORMIC ACID | | Authors: | Liu, L, Zhang, S.Q. | | Deposit date: | 2018-01-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
6C4Z
 
 | |
6C52
 
 | | Cross-alpha Amyloid-like Structure alphaTet | | Descriptor: | Cross-alpha Amyloid-like Structure alphaTet, GLYCEROL | | Authors: | Liu, L, Zhang, S.Q. | | Deposit date: | 2018-01-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
6C51
 
 | | Cross-alpha Amyloid-like Structure alphaAmL | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cross-alpha Amyloid-like Structure alphaAmL, PHOSPHATE ION | | Authors: | Liu, L, Zhang, S.Q. | | Deposit date: | 2018-01-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
4YL9
 
 | | Crystal Structure of wild-type of hsp14.1 from Sulfolobus solfatataricus P2 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Heat shock protein Hsp20 | | Authors: | Liu, L, Chen, J.Y, Yun, C.H. | | Deposit date: | 2015-03-05 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Active-State Structures of a Small Heat-Shock Protein Revealed a Molecular Switch for Chaperone Function
Structure, 23, 2015
|
|
2B3R
 
 | | Crystal structure of the C2 domain of class II phosphatidylinositide 3-kinase C2 | | Descriptor: | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide, SULFATE ION | | Authors: | Liu, L, Song, X, He, D, Komma, C, Kita, A, Verbasius, J.V, Bellamy, H, Miki, K, Czech, M.P, Zhou, G.W. | | Deposit date: | 2005-09-20 | | Release date: | 2005-12-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the C2 domain of class II phosphatidylinositide 3-kinase C2alpha.
J.Biol.Chem., 281, 2006
|
|
5XWP
 
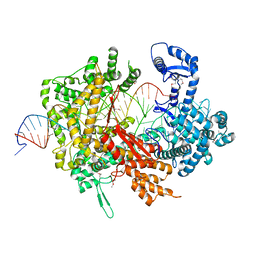 | | Crystal structure of LbuCas13a-crRNA-target RNA ternary complex | | Descriptor: | RNA (30-MER), RNA (59-MER), Uncharacterized protein | | Authors: | Liu, L, Li, X, Li, Z, Wang, Y. | | Deposit date: | 2017-06-30 | | Release date: | 2017-09-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.086 Å) | | Cite: | The Molecular Architecture for RNA-Guided RNA Cleavage by Cas13a.
Cell, 170, 2017
|
|
5WYB
 
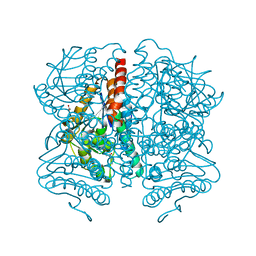 | | Structure of Pseudomonas aeruginosa DspI | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ACETATE ION, Probable enoyl-CoA hydratase/isomerase | | Authors: | Liu, L, Peng, C, Li, T, Li, C, He, L, Song, Y, Zhu, Y, Shen, Y, Bao, R. | | Deposit date: | 2017-01-12 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional studies on Pseudomonas aeruginosa DspI: implications for its role in DSF biosynthesis.
Sci Rep, 8, 2018
|
|
2PL5
 
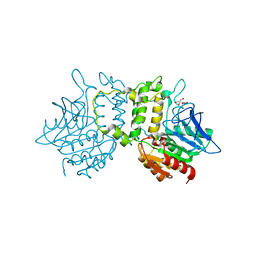 | | Crystal Structure of Homoserine O-acetyltransferase from Leptospira interrogans | | Descriptor: | GLYCEROL, Homoserine O-acetyltransferase | | Authors: | Liu, L, Wang, M, Wang, Y, Wei, Z, Xu, H, Gong, W. | | Deposit date: | 2007-04-19 | | Release date: | 2007-11-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of homoserine O-acetyltransferase from Leptospira interrogans
Biochem.Biophys.Res.Commun., 363, 2007
|
|
3GUP
 
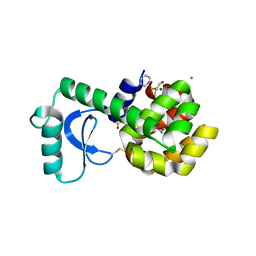 | |
3GUN
 
 | |
