6PEC
 
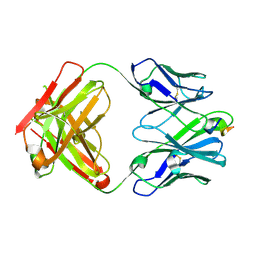 | |
6PDS
 
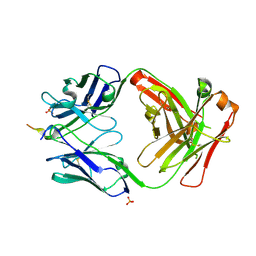 | | Vaccine-elicited NHP FP-targeting antibody 0PV-a.04 in complex with HIV fusion peptide (residue 512-519) | | Descriptor: | HIV-1 fusion peptide residue 512-519, SULFATE ION, antibody 0PV-a.04 heavy chain, ... | | Authors: | Xu, K, Liu, K, Wang, Y, Kwong, P.D. | | Deposit date: | 2019-06-19 | | Release date: | 2020-06-24 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Modular recognition of antigens provides a mechanism that improves vaccine-elicited antibody-class frequencies
To Be Published
|
|
6PDR
 
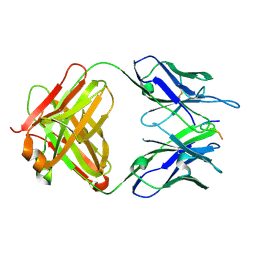 | |
6PDU
 
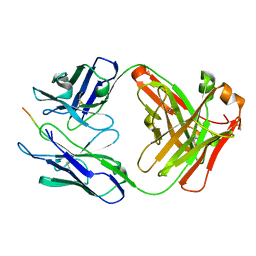 | | Vaccine-elicited NHP FP-targeting antibody 13N024-a.01 in complex with HIV fusion peptide (residue 512-519) | | Descriptor: | HIV-1 fusion peptide residue 512-519, antibody 13N024-a.01, heavy chain, ... | | Authors: | Xu, K, Liu, K, Wang, Y, Kwong, P.D. | | Deposit date: | 2019-06-19 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Modular recognition of antigens provides a mechanism that improves vaccine-elicited antibody-class frequencies
To Be Published
|
|
5VEI
 
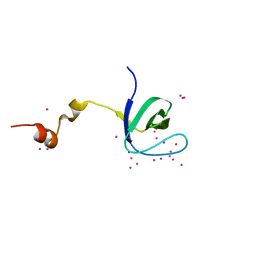 | | Crystal structure of the SH3 domain of human sorbin and SH3 domain-containing protein 2 | | Descriptor: | Sorbin and SH3 domain-containing protein 2, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Huang, H, Gu, J, Liu, K, Sidhu, S.S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-04-04 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Crystal structure of the SH3 domain of human sorbin and SH3 domain-containing protein 2
To be Published
|
|
8K3D
 
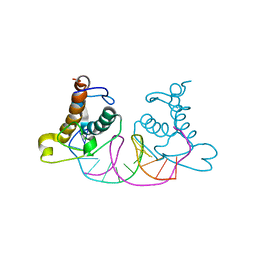 | | Crystal structure of NRF1 DBD bound to DNA | | Descriptor: | DNA (5'-D(*GP*GP*TP*GP*CP*GP*CP*AP*TP*GP*CP*GP*CP*AP*CP*C)-3'), Nuclear respiratory factor 1 | | Authors: | Li, W.F, Liu, K, Min, J.R. | | Deposit date: | 2023-07-15 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanism of specific DNA sequence recognition by NRF1.
Nucleic Acids Res., 52, 2024
|
|
4RCI
 
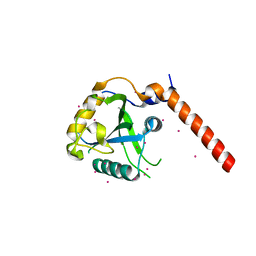 | | Crystal structure of YTHDF1 YTH domain | | Descriptor: | UNKNOWN ATOM OR ION, YTH domain-containing family protein 1 | | Authors: | Xu, C, Tempel, W, Liu, K, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-09-16 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis for the Discriminative Recognition of N6-Methyladenosine RNA by the Human YT521-B Homology Domain Family of Proteins.
J.Biol.Chem., 290, 2015
|
|
4R3I
 
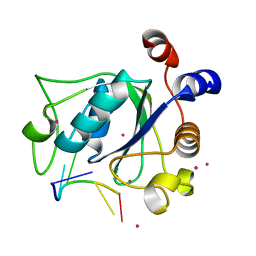 | | The crystal structure of an RNA complex | | Descriptor: | RNA (5'-R(*GP*GP*(6MZ)P*CP*U)-3'), UNKNOWN ATOM OR ION, YTH domain-containing protein 1 | | Authors: | Tempel, W, Xu, C, Liu, K, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-08-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for selective binding of m(6)A RNA by the YTHDC1 YTH domain.
Nat.Chem.Biol., 10, 2014
|
|
4UPF
 
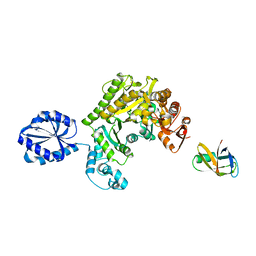 | | Assembly principles of the unique cage formed by the ATPase RavA hexamer and the lysine decarboxylase LdcI decamer | | Descriptor: | ATPASE RAVA, LYSINE DECARBOXYLASE, INDUCIBLE | | Authors: | Malet, H, Liu, K, El Bakkouri, M, Chan, S.W.S, Effantin, G, Bacia, M, Houry, W.A, Gutsche, I. | | Deposit date: | 2014-06-16 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Assembly Principles of a Unique Cage Formed by Hexameric and Decameric E. Coli Proteins.
Elife, 3, 2014
|
|
4UPB
 
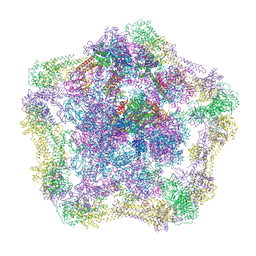 | | Electron cryo-microscopy of the complex formed between the hexameric ATPase RavA and the decameric inducible decarboxylase LdcI | | Descriptor: | ATPASE RAVA, LYSINE DECARBOXYLASE, INDUCIBLE | | Authors: | Malet, H, Liu, K, El Bakkouri, M, Chan, S.W.S, Effantin, G, Bacia, M, Houry, W.A, Gutsche, I. | | Deposit date: | 2014-06-15 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Assembly Principles of a Unique Cage Formed by Hexameric and Decameric E. Coli Proteins.
Elife, 3, 2014
|
|
7YIM
 
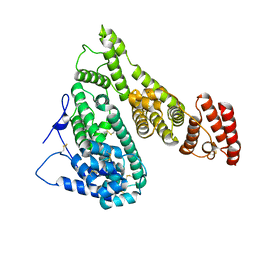 | | Cryo-EM structure of human Alpha-fetoprotein | | Descriptor: | Alpha-fetoprotein | | Authors: | Liu, N, Liu, K, Wu, C, Liu, Z, Li, M, Wang, J, Wang, H.W. | | Deposit date: | 2022-07-17 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Uniform thin ice on ultraflat graphene for high-resolution cryo-EM.
Nat.Methods, 20, 2023
|
|
1D3R
 
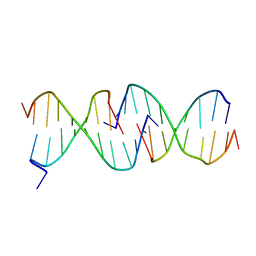 | | CRYSTAL STRUCTURE OF TRIPLEX DNA | | Descriptor: | DNA (5'-D(*CP*(BRU)P*CP*CP*(BRU)P*CP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*CP*GP*GP*AP*G)-3') | | Authors: | Rhee, S, Han, Z.-J, Liu, K, Todd Miles, H.T, Davies, D.R. | | Deposit date: | 1999-09-30 | | Release date: | 2000-01-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a triple helical DNA with a triplex-duplex junction.
Biochemistry, 38, 1999
|
|
1JUU
 
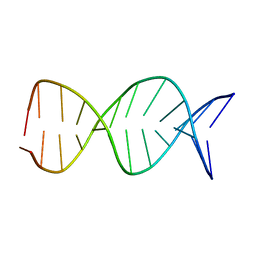 | | NMR Structure of a Parallel Stranded DNA Duplex at Atomic Resolution | | Descriptor: | 5'-D(P*CP*CP*AP*TP*AP*AP*TP*TP*TP*AP*CP*C)-3', 5'-D(P*CP*CP*TP*AP*TP*TP*AP*AP*AP*TP*CP*C)-3' | | Authors: | Parvathy, V.R, Bhaumik, S.R, Chary, K.V.R, Govil, G, Liu, K, Howard, F.B, Miles, H.T. | | Deposit date: | 2001-08-28 | | Release date: | 2002-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a parallel-stranded DNA duplex at atomic resolution.
Nucleic Acids Res., 30, 2002
|
|
8K8A
 
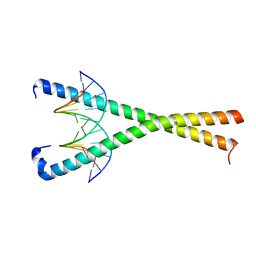 | | Crystal structure of NFIL3 in complex with TTACGTAA DNA | | Descriptor: | DNA (5'-D(*CP*AP*TP*TP*AP*CP*GP*TP*AP*AP*TP*G)-3'), Nuclear factor interleukin-3-regulated protein | | Authors: | Min, J.R, Chen, S.Z, Liu, K. | | Deposit date: | 2023-07-29 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis for specific DNA sequence recognition by the transcription factor NFIL3.
J.Biol.Chem., 300, 2024
|
|
8K89
 
 | | Crystal structure of NFIL3 | | Descriptor: | Nuclear factor interleukin-3-regulated protein | | Authors: | Min, J.R, Chen, S.Z, Liu, K. | | Deposit date: | 2023-07-29 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for specific DNA sequence recognition by the transcription factor NFIL3.
J.Biol.Chem., 300, 2024
|
|
8K8D
 
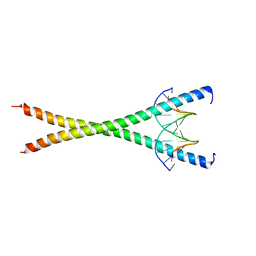 | |
8K86
 
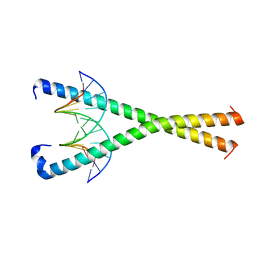 | | Crystal structure of NFIL3 in complex with TTATGTAA DNA | | Descriptor: | DNA (5'-D(*CP*AP*TP*TP*AP*TP*GP*TP*AP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*TP*AP*CP*AP*TP*AP*AP*TP*G)-3'), Nuclear factor interleukin-3-regulated protein | | Authors: | Min, J.R, Chen, S.Z, Liu, K. | | Deposit date: | 2023-07-28 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for specific DNA sequence recognition by the transcription factor NFIL3.
J.Biol.Chem., 300, 2024
|
|
8K8C
 
 | | Crystal structure of C/EBPalpha BZIP domain bound to a high affinity DNA | | Descriptor: | CCAAT/enhancer-binding protein alpha, DNA (5'-D(*CP*AP*TP*TP*AP*CP*GP*TP*AP*AP*TP*GP*A)-3'), DNA (5'-D(*CP*AP*TP*TP*AP*CP*GP*TP*AP*AP*TP*GP*T)-3'), ... | | Authors: | Min, J.R, Chen, S.Z, Liu, K. | | Deposit date: | 2023-07-29 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for specific DNA sequence recognition by the transcription factor NFIL3.
J.Biol.Chem., 300, 2024
|
|
5TKK
 
 | |
5TKJ
 
 | | Structure of vaccine-elicited diverse HIV-1 neutralizing antibody vFP1.01 in complex with HIV-1 fusion peptide residue 512-519 | | Descriptor: | HIV-1 fusion peptide residue 512-519, SULFATE ION, vFP1.01 chimeric mouse antibody heavy chain, ... | | Authors: | Xu, K, Liu, K, Kwong, P.D. | | Deposit date: | 2016-10-06 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.118 Å) | | Cite: | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
6CDI
 
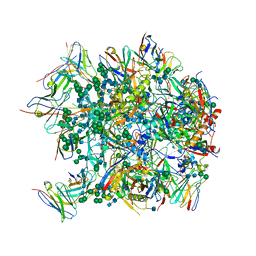 | | Cryo-EM structure at 3.6 A resolution of vaccine-elicited antibody vFP16.02 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein 120, ... | | Authors: | Acharya, P, Xu, K, Liu, K, Carragher, B, Potter, C.S, Kwong, P.D. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-16 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
6CDM
 
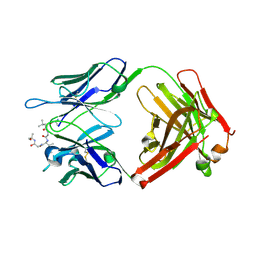 | |
6CDO
 
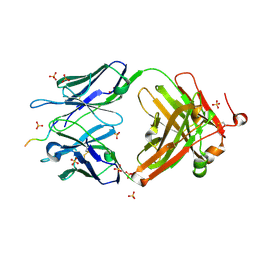 | | Structure of vaccine-elicited HIV-1 neutralizing antibody vFP16.02 in complex with HIV-1 fusion peptide residue 512-519 | | Descriptor: | HIV-1 fusion peptide 512-519, SULFATE ION, vFP16.02 Fab heavy chain, ... | | Authors: | Xu, K, Liu, K, Kwong, P.D. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-16 | | Last modified: | 2018-06-20 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
6CDP
 
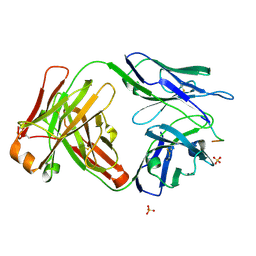 | | Vaccine-elicited HIV-1 neutralizing antibody vFP20.01 in complex with HIV-1 fusion peptide residue 512-519 | | Descriptor: | HIV-1 fusion peptide 512-519, SULFATE ION, vFP20.01 Fab heavy chain, ... | | Authors: | Xu, K, Liu, K, Kwong, P.D. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-16 | | Last modified: | 2018-06-20 | | Method: | X-RAY DIFFRACTION (2.456 Å) | | Cite: | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
8X1N
 
 | | Cryo-EM structure of human alpha-fetoprotein | | Descriptor: | Alpha-fetoprotein, PALMITIC ACID, ZINC ION, ... | | Authors: | Liu, Z.M, Li, M.S, Wu, C, Liu, K. | | Deposit date: | 2023-11-08 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structural characteristics of alpha-fetoprotein, including N-glycosylation, metal ion and fatty acid binding sites.
Commun Biol, 7, 2024
|
|
