2B9Z
 
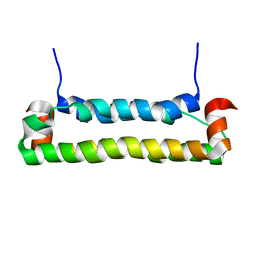 | | Solution structure of FHV B2, a viral suppressor of RNAi | | Descriptor: | B2 protein | | Authors: | Lingel, A, Simon, B, Izaurralde, E, Sattler, M. | | Deposit date: | 2005-10-13 | | Release date: | 2005-11-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the flock house virus B2 protein, a viral suppressor of RNA interference, shows a novel mode of double-stranded RNA recognition.
Embo Rep., 6, 2005
|
|
1T2R
 
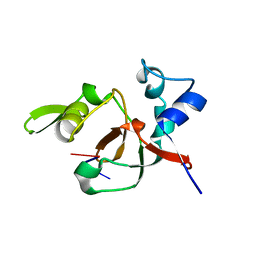 | | Structural basis for 3' end recognition of nucleic acids by the Drosophila Argonaute 2 PAZ domain | | Descriptor: | 5'-R(*CP*UP*CP*AP*C)-3', Argonaute 2 | | Authors: | Lingel, A, Simon, B, Izaurralde, E, Sattler, M. | | Deposit date: | 2004-04-22 | | Release date: | 2004-06-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nucleic acid 3'-end recognition by the Argonaute2 PAZ domain.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1T2S
 
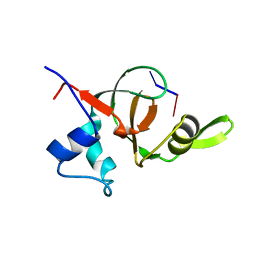 | | Structural basis for 3' end recognition of nucleic acids by the Drosophila Argonaute 2 PAZ domain | | Descriptor: | 5'-D(*CP*TP*CP*AP*C)-3', Argonaute 2 | | Authors: | Lingel, A, Simon, B, Izaurralde, E, Sattler, M. | | Deposit date: | 2004-04-22 | | Release date: | 2004-06-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nucleic acid 3'-end recognition by the Argonaute2 PAZ domain.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1VYN
 
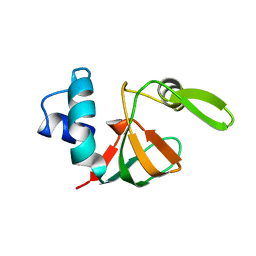 | |
2KDD
 
 | |
2KLL
 
 | | Solution structure of human interleukin-33 | | Descriptor: | Interleukin-33 | | Authors: | Lingel, A, Fairbrother, W. | | Deposit date: | 2009-07-06 | | Release date: | 2009-11-17 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of IL-33 and its interaction with the ST2 and IL-1RAcP receptors--insight into heterotrimeric IL-1 signaling complexes.
Structure, 17, 2009
|
|
2VXG
 
 | | Crystal structure of the conserved C-terminal region of Ge-1 | | Descriptor: | CG6181-PA, ISOFORM A | | Authors: | Jinek, M, Eulalio, A, Lingel, A, Helms, S, Conti, E, Izaurralde, E. | | Deposit date: | 2008-07-04 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The C-Terminal Region of Ge-1 Presents Conserved Structural Features Required for P-Body Localization.
RNA, 14, 2008
|
|
3T6P
 
 | | IAP antagonist-induced conformational change in cIAP1 promotes E3 ligase activation via dimerization | | Descriptor: | Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Dueber, E.C, Schoeffler, A.J, Lingel, A, Elliott, M, Fedorova, A.V, Giannetti, A.M, Zobel, K, Maurer, B, Varfolomeev, E, Wu, P, Wallweber, H, Hymowitz, S, Deshayes, K, Vucic, D, Fairbrother, W.J. | | Deposit date: | 2011-07-28 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Antagonists induce a conformational change in cIAP1 that promotes autoubiquitination.
Science, 334, 2011
|
|
2KJX
 
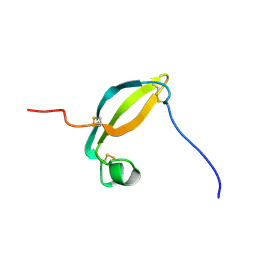 | | Solution structure of the extracellular domain of JTB | | Descriptor: | Jumping translocation breakpoint protein | | Authors: | Rousseau, F, Lingel, A, Pan, B, Fairbrother, W.J, Bazan, F. | | Deposit date: | 2009-06-10 | | Release date: | 2010-08-11 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The structure of the extracellular domain of the jumping translocation breakpoint protein reveals a variation of the midkine fold.
J.Mol.Biol., 415, 2012
|
|
6B7D
 
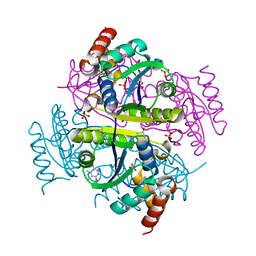 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with 3-(4-chlorophenyl)-6-methoxy-4,5-dimethylpyridazine | | Descriptor: | 3-(4-chlorophenyl)-6-methoxy-4,5-dimethylpyridazine, DIMETHYL SULFOXIDE, POTASSIUM ION, ... | | Authors: | Proudfoot, A.W, Bussiere, D, Lingel, A. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-Confidence Protein-Ligand Complex Modeling by NMR-Guided Docking Enables Early Hit Optimization.
J. Am. Chem. Soc., 139, 2017
|
|
6B7A
 
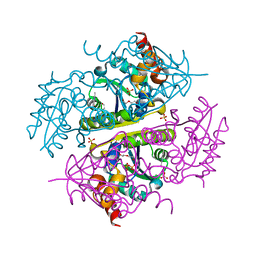 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with 2-methyl-1H-benzo[d]imidazol-4-ol | | Descriptor: | 2-methyl-1H-benzimidazol-7-ol, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Proudfoot, A.W, Bussiere, D, Lingel, A. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | High-Confidence Protein-Ligand Complex Modeling by NMR-Guided Docking Enables Early Hit Optimization.
J. Am. Chem. Soc., 139, 2017
|
|
6B7C
 
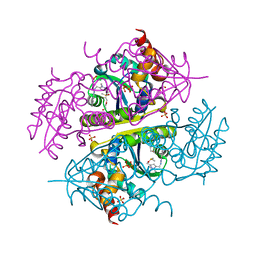 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with N-((1,3-dimethyl-1H-pyrazol-5-yl)methyl)-5-methyl-1H-imidazo[4,5-b]pyridin-2-amine | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, N-[(1,3-dimethyl-1H-pyrazol-5-yl)methyl]-5-methyl-3H-imidazo[4,5-b]pyridin-2-amine, ... | | Authors: | Proudfoot, A.W, Bussiere, D, Lingel, A. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.564 Å) | | Cite: | High-Confidence Protein-Ligand Complex Modeling by NMR-Guided Docking Enables Early Hit Optimization.
J. Am. Chem. Soc., 139, 2017
|
|
6B7E
 
 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with (R)-4-(5-(difluoromethyl)-1H-imidazol-1-yl)-3,3-dimethylisochroman-1-one | | Descriptor: | (4R)-4-[5-(difluoromethyl)-1H-imidazol-1-yl]-3,3-dimethyl-3,4-dihydro-1H-2-benzopyran-1-one, DIMETHYL SULFOXIDE, Phosphopantetheine adenylyltransferase, ... | | Authors: | Proudfoot, A.W, Bussiere, D, Lingel, A. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | High-Confidence Protein-Ligand Complex Modeling by NMR-Guided Docking Enables Early Hit Optimization.
J. Am. Chem. Soc., 139, 2017
|
|
6B7F
 
 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with (R)-3,3-dimethyl-4-(5-vinyl-1H-imidazol-1-yl)isochroman-1-one | | Descriptor: | (4R)-4-(5-ethenyl-1H-imidazol-1-yl)-3,3-dimethyl-3,4-dihydro-1H-2-benzopyran-1-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Proudfoot, A.W, Bussiere, D, Lingel, A. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.562 Å) | | Cite: | High-Confidence Protein-Ligand Complex Modeling by NMR-Guided Docking Enables Early Hit Optimization.
J. Am. Chem. Soc., 139, 2017
|
|
6B7B
 
 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with 5-methoxy-2-methyl-1H-indole | | Descriptor: | 5-methoxy-2-methyl-1H-indole, DIMETHYL SULFOXIDE, PYROPHOSPHATE 2-, ... | | Authors: | Proudfoot, A.W, Bussiere, D, Lingel, A. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | High-Confidence Protein-Ligand Complex Modeling by NMR-Guided Docking Enables Early Hit Optimization.
J. Am. Chem. Soc., 139, 2017
|
|
