315D
 
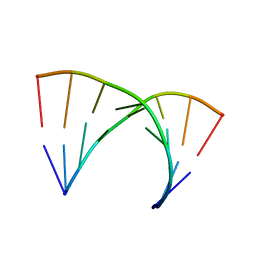 | |
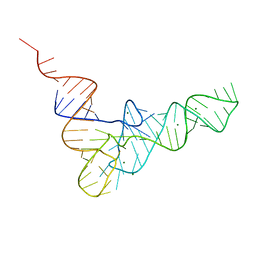
6EDI
 
 | |
1TRA
 
 | | RESTRAINED REFINEMENT OF THE MONOCLINIC FORM OF YEAST PHENYLALANINE TRANSFER RNA. TEMPERATURE FACTORS AND DYNAMICS, COORDINATED WATERS, AND BASE-PAIR PROPELLER TWIST ANGLES | | Descriptor: | MAGNESIUM ION, TRNAPHE | | Authors: | Westhof, E, Sundaralingam, M. | | Deposit date: | 1986-05-16 | | Release date: | 1986-07-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Restrained refinement of the monoclinic form of yeast phenylalanine transfer RNA. Temperature factors and dynamics, coordinated waters, and base-pair propeller twist angles.
Biochemistry, 25, 1986
|
|
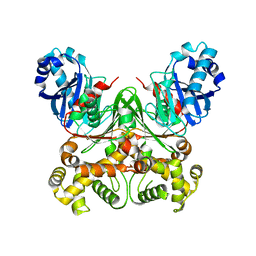
5ARC
 
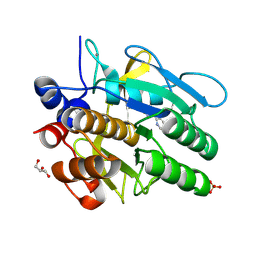 | | Cooperative bio-metallic selectivity in a tailored protease enables creation of a C-C cross-coupling Heckase | | Descriptor: | 5-methyl-2-(5-methylpyridin-2-yl)pyridine, CALCIUM ION, GLYCEROL, ... | | Authors: | Sharma, M, Diaz-Rodriguez, A, Offen, W.A, Palm-Espling, M.E, Pordea, A, Wormald, M.R, Mcdonough, M, Davies, G.J, Davis, B.G. | | Deposit date: | 2015-09-24 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Cooperative Bio-Metallic Selectivity in a Tailored Protease Enables Creation of a C-C Cross-Coupling Heckase
To be Published
|
|
1D79
 
 | | HIGH RESOLUTION REFINEMENT OF THE HEXAGONAL A-DNA OCTAMER D(GTGTACAC) AT 1.4 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*GP*TP*GP*TP*AP*CP*AP*C)-3') | | Authors: | Thota, N, Li, X.H, Bingman, C.A, Sundaralingam, M. | | Deposit date: | 1992-06-12 | | Release date: | 1993-04-15 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution refinement of the hexagonal A-DNA octamer d(GTGTACAC) at 1.4 A.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
304D
 
 | |
5ARB
 
 | | Cooperative bio-metallic selectivity in a tailored protease enables creation of a C-C cross-coupling Heckase | | Descriptor: | 5-methyl-2-(5-methylpyridin-2-yl)pyridine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sharma, M, Diaz-Rodriguez, A, Offen, W.A, Palm-Espling, M.E, Pordea, A, Wormald, M.R, Mcdonough, M, Davies, G.J, Davis, B.G. | | Deposit date: | 2015-09-24 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Cooperative Bio-Metallic Selectivity in a Tailored Protease Enables Creation of a C-C Cross-Coupling Heckase
To be Published
|
|
5ARD
 
 | | Cooperative bio-metallic selectivity in a tailored protease enables creation of a C-C cross-coupling Heckase | | Descriptor: | 5-methyl-2-(5-methylpyridin-2-yl)pyridine, CALCIUM ION, GLYCEROL, ... | | Authors: | Sharma, M, Diaz-Rodriguez, A, Offen, W.A, Palm-Espling, M.E, Pordea, A, Wormald, M.R, Mcdonough, M, Davies, G.J, Davis, B.G. | | Deposit date: | 2015-09-24 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Cooperative Bio-Metallic Selectivity in a Tailored Protease Enables Creation of a C-C Cross-Coupling Heckase
To be Published
|
|
323D
 
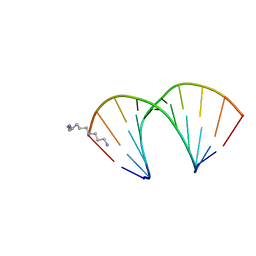 | | CRYSTAL STRUCTURES OF D(CCGGGCCM5CGG)-ORTHOGONAL FORM | | Descriptor: | DNA (5'-D(*CP*CP*GP*GP*GP*CP*CP*(5CM)P*GP*G)-3'), SPERMINE | | Authors: | Tippin, D.B, Sundaralingam, M. | | Deposit date: | 1997-03-17 | | Release date: | 1997-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Nine polymorphic crystal structures of d(CCGGGCCCGG), d(CCGGGCCm5CGG), d(Cm5CGGGCCm5CGG) and d(CCGGGCC(Br)5CGG) in three different conformations: effects of spermine binding and methylation on the bending and condensation of A-DNA.
J.Mol.Biol., 267, 1997
|
|
395D
 
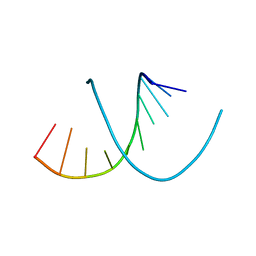 | |
325D
 
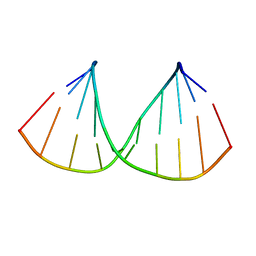 | | CRYSTAL STRUCTURES OF D(CM5CGGGCCM5CGG)-HEXAGONAL FORM | | Descriptor: | DNA (5'-D(*CP*(5CM)P*GP*GP*GP*CP*CP*(5CM)P*GP*G)-3') | | Authors: | Tippin, D.B, Sundaralingam, M. | | Deposit date: | 1997-03-17 | | Release date: | 1997-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nine polymorphic crystal structures of d(CCGGGCCCGG), d(CCGGGCCm5CGG), d(Cm5CGGGCCm5CGG) and d(CCGGGCC(Br)5CGG) in three different conformations: effects of spermine binding and methylation on the bending and condensation of A-DNA.
J.Mol.Biol., 267, 1997
|
|
375D
 
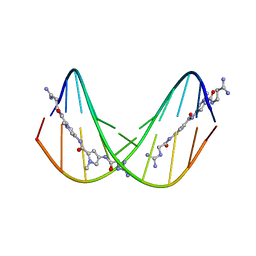 | | A NOVEL END-TO-END BINDING OF TWO NETROPSINS TO THE DNA DECAMER D(CCCCCIIIII)2 | | Descriptor: | DNA (5'-D(*CP*CP*CP*CP*CP*IP*IP*IP*IP*I)-3'), NETROPSIN | | Authors: | Chen, X, Rao, S.T, Sekar, K, Sundaralingam, M. | | Deposit date: | 1998-01-14 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Novel End-to-End Binding of Two Netropsins to the DNA Decamers d(CCCCCIIIII) 2, d(CCCBr5CCIIIII)2, d(CBr5CCCCIIIII)2
Nucleic Acids Res., 26, 1998
|
|
8QHH
 
 | |
8QHI
 
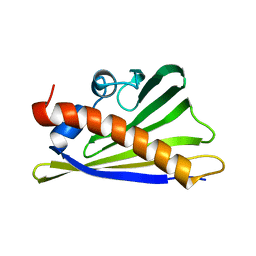 | |
5AQ8
 
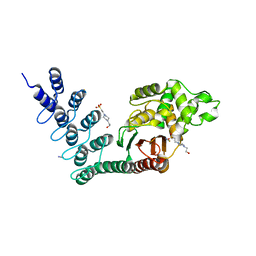 | | DARPin-based Crystallization Chaperones exploit Molecular Geometry as a Screening Dimension in Protein Crystallography | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, OFF7_DB12V4, THIOCYANATE ION | | Authors: | Batyuk, A, Wu, Y, Honegger, A, Heberling, M, Plueckthun, A. | | Deposit date: | 2015-09-21 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Darpin-Based Crystallization Chaperones Exploit Molecular Geometry as a Screening Dimension in Protein Crystallography
J.Mol.Biol., 428, 2016
|
|
5AQA
 
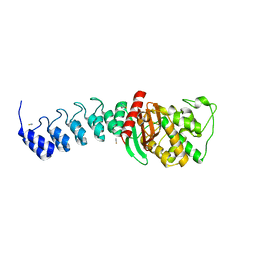 | | DARPin-based Crystallization Chaperones exploit Molecular Geometry as a Screening Dimension in Protein Crystallography | | Descriptor: | OFF7_DB04V3, THIOCYANATE ION | | Authors: | Batyuk, A, Wu, Y, Honegger, A, Heberling, M, Plueckthun, A. | | Deposit date: | 2015-09-21 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Darpin-Based Crystallization Chaperones Exploit Molecular Geometry as a Screening Dimension in Protein Crystallography
J.Mol.Biol., 428, 2016
|
|
5AQ7
 
 | | DARPin-based Crystallization Chaperones exploit Molecular Geometry as a Screening Dimension in Protein Crystallography | | Descriptor: | D12_DB04V3, MALONATE ION | | Authors: | Batyuk, A, Wu, Y, Honegger, A, Heberling, M, Plueckthun, A. | | Deposit date: | 2015-09-21 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Darpin-Based Crystallization Chaperones Exploit Molecular Geometry as a Screening Dimension in Protein Crystallography
J.Mol.Biol., 428, 2016
|
|
5AQE
 
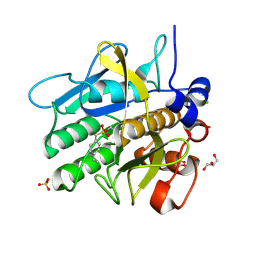 | | Cooperative bio-metallic selectivity in a tailored protease enables creation of a C-C cross-coupling Heckase | | Descriptor: | (4-VINYLPHENYL)METHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sharma, M, Diaz-Rodriguez, A, Offen, W.A, Palm-Espling, M.E, Pordea, A, Wormald, M.R, Mcdonough, M, Davies, G.J, Davis, B.G. | | Deposit date: | 2015-09-22 | | Release date: | 2015-10-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Bisubstrate Localisation by a Tailored Serine Protease Allows Creation of a Heck-Ase
To be Published
|
|
4N71
 
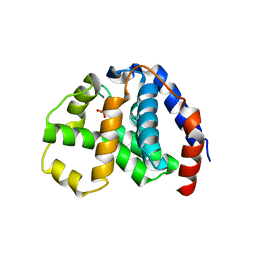 | | X-Ray Crystal Structure of 2-amino-1-hydroxyethylphosphonate-bound PhnZ | | Descriptor: | FE (III) ION, Predicted HD phosphohydrolase PhnZ, [(1R)-2-amino-1-hydroxyethyl]phosphonic acid | | Authors: | Woersdoerfer, B, Lingaraju, M, Yennawar, N, Boal, A.K, Krebs, C, Bollinger Jr, J.M, Pandelia, M.-E. | | Deposit date: | 2013-10-14 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.984 Å) | | Cite: | Organophosphonate-degrading PhnZ reveals an emerging family of HD domain mixed-valent diiron oxygenases.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4N6W
 
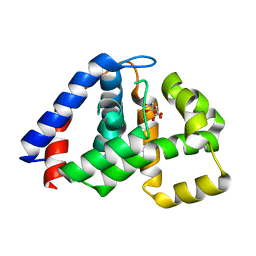 | | X-Ray Crystal Structure of Citrate-bound PhnZ | | Descriptor: | CITRATE ANION, FE (III) ION, Predicted HD phosphohydrolase PhnZ | | Authors: | Worsdorfer, B, Lingaraju, M, Yennawar, N.H, Boal, A.K, Krebs, C, Bollinger Jr, J.M, Pandelia, M.E. | | Deposit date: | 2013-10-14 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Organophosphonate-degrading PhnZ reveals an emerging family of HD domain mixed-valent diiron oxygenases.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8U6Y
 
 | | Preholo-Proteasome from Beta 3 D205 deletion | | Descriptor: | Proteasome assembly chaperone 2, Proteasome chaperone 1, Proteasome maturation factor UMP1, ... | | Authors: | Walsh Jr, R.M, Rawson, S, Velez, B, Blickling, M, Razi, A, Hanna, J. | | Deposit date: | 2023-09-14 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of autocatalytic activation during proteasome assembly.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8U7U
 
 | | Proteasome 20S Core Particle from Beta 3 D205 deletion | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Walsh Jr, R.M, Rawson, S, Velez, B, Blickling, M, Razi, A, Hanna, J. | | Deposit date: | 2023-09-15 | | Release date: | 2024-04-17 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.16 Å) | | Cite: | Mechanism of autocatalytic activation during proteasome assembly.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6N82
 
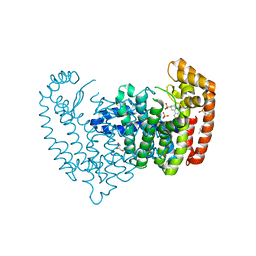 | | Crystal structure of human FPPS in complex with an allosteric inhibitor YF-02037 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Farnesyl pyrophosphate synthase, ... | | Authors: | Park, J, Schilling, M.A, Berghuis, A.M. | | Deposit date: | 2018-11-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chirality-Driven Mode of Binding of alpha-Aminophosphonic Acid-Based Allosteric Inhibitors of the Human Farnesyl Pyrophosphate Synthase (hFPPS).
J.Med.Chem., 62, 2019
|
|
6N83
 
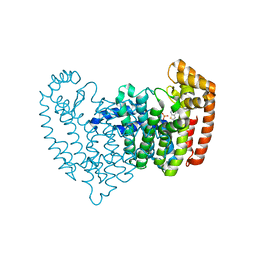 | | Crystal structure of human FPPS in complex with an allosteric inhibitor YF-02037 | | Descriptor: | CHLORIDE ION, Farnesyl pyrophosphate synthase, PHOSPHATE ION, ... | | Authors: | Park, J, Schilling, M.A, Berghuis, A.M. | | Deposit date: | 2018-11-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chirality-Driven Mode of Binding of alpha-Aminophosphonic Acid-Based Allosteric Inhibitors of the Human Farnesyl Pyrophosphate Synthase (hFPPS).
J.Med.Chem., 62, 2019
|
|
8B1U
 
 | | RecBCD-DNA in complex with the phage protein Abc2 and host PpiB | | Descriptor: | Anti-RecBCD protein 2, DNA (70-MER), MAGNESIUM ION, ... | | Authors: | Wilkinson, M, Wilkinson, O.J, Feyerherm, C, Fletcher, E.E, Wigley, D.B, Dillingham, M.S. | | Deposit date: | 2022-09-12 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of RecBCD in complex with phage-encoded inhibitor proteins reveal distinctive strategies for evasion of a bacterial immunity hub.
Elife, 11, 2022
|
|
