6P58
 
 | | Dark and Steady State-Illuminated Crystal Structure of Cyanobacteriochrome Receptor PixJ at 150K | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Methyl-accepting chemotaxis protein, ... | | Authors: | Clinger, J.A, Miller, M.D, Buirgie, E.S, Vierstra, R.D, Phillips Jr, G.N. | | Deposit date: | 2019-05-29 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Photoreversible interconversion of a phytochrome photosensory module in the crystalline state.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6N04
 
 | | The X-ray crystal structure of AbsH3, an FAD dependent reductase from the Abyssomicin biosynthesis pathway in Streptomyces | | Descriptor: | AbsH3, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Clinger, J.A, Wang, X, Cai, W, Miller, M.D, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-11-06 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | The crystal structure of AbsH3: A putative flavin adenine dinucleotide-dependent reductase in the abyssomicin biosynthesis pathway.
Proteins, 2020
|
|
6ND7
 
 | | The crystal structure of TerB co-crystallized with polyporic acid | | Descriptor: | 2~3~,2~6~-dihydroxy[1~1~,2~1~:2~4~,3~1~-terphenyl]-2~2~,2~5~-dione, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Clinger, J.A, Elshahawi, S.I, Zhang, Y, Hall, R.P, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2018-12-13 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structure and Function of Terfestatin Biosynthesis Enzymes TerB and TerC
To Be Published
|
|
8E20
 
 | |
8EDM
 
 | |
6MGO
 
 | |
6D2V
 
 | | Apo Structure of TerB, an NADP Dependent Oxidoreductase in the Terfestatin Biosynthesis Pathway | | Descriptor: | CHLORIDE ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, THIOCYANATE ION, ... | | Authors: | Clinger, J.A, Elshahawi, S.I, Zhang, Y, Hall, R.P, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2018-04-14 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structure and Function of Terfestatin Biosynthesis Enzymes TerB and TerC
To Be Published
|
|
7L36
 
 | | PEPCK steady-state structure with Mn and GTP | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Clinger, J.A, Moreau, D.W, McLeod, M.J, Holyoak, T, Thorne, R.E. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Millisecond mix-and-quench crystallography (MMQX) enables time-resolved studies of PEPCK with remote data collection.
Iucrj, 8, 2021
|
|
7L3M
 
 | | PEPCK MMQX structure 40ms post-mixing with oxaloacetic acid | | Descriptor: | CARBON DIOXIDE, GUANOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Clinger, J.A, Moreau, D.W, McLeod, M.J, Holyoak, T, Thorne, R.E. | | Deposit date: | 2020-12-18 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Millisecond mix-and-quench crystallography (MMQX) enables time-resolved studies of PEPCK with remote data collection.
Iucrj, 8, 2021
|
|
7L3V
 
 | | PEPCK MMQX structure 120ms post-mixing with oxaloacetic acid | | Descriptor: | CARBON DIOXIDE, GUANOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Clinger, J.A, Moreau, D.W, McLeod, M.J, Holyoak, T, Thorne, R.E. | | Deposit date: | 2020-12-18 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Millisecond mix-and-quench crystallography (MMQX) enables time-resolved studies of PEPCK with remote data collection.
Iucrj, 8, 2021
|
|
8EDO
 
 | |
8EDN
 
 | |
8EDL
 
 | |
3HWC
 
 | |
6D34
 
 | | Apo Crystal Structure of TerC, a Terfestatin Biosynthesis Enzyme | | Descriptor: | ISOPROPYL ALCOHOL, TerC | | Authors: | Clinger, J.A, Elshahawi, S.I, Zhang, Y, Hall, R.P, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2018-04-14 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Function of Terfestatin Biosynthesis Enzymes TerB and TerC
To Be Published
|
|
4K5A
 
 | | Co-crystallization with conformation-specific designed ankyrin repeat proteins explains the conformational flexibility of BCL-W | | Descriptor: | Bcl-2-like protein 2, Designed Ankyrin Repeat Protein 013_D12 | | Authors: | Schilling, J, Schoeppe, J, Sauer, E, Plueckthun, A. | | Deposit date: | 2013-04-14 | | Release date: | 2014-04-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Co-Crystallization with Conformation-Specific Designed Ankyrin Repeat Proteins Explains the Conformational Flexibility of BCL-W
J.Mol.Biol., 426, 2014
|
|
3M6G
 
 | | Crystal structure of actin in complex with lobophorolide | | Descriptor: | (1S,3S,4S,5S,7R,8S,9R,12E,14E,16R,17R,19R)-16-hydroxy-9-{(1S,2S,3S)-2-hydroxy-5-[(2S,4R,6S)-4-methoxy-6-methyltetrahydro-2H-pyran-2-yl]-1,3-dimethylpentyl}-3,5,7,17-tetramethoxy-8,14-dimethyl-11H-spiro[10,23-dioxabicyclo[17.3.1]tricosa-12,14,20-triene-4,2'-oxiran]-11-one, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Allingham, J.S. | | Deposit date: | 2010-03-15 | | Release date: | 2010-09-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two molecules of lobophorolide cooperate to stabilize an actin dimer using both their "ring" and "tail" region.
Chem.Biol., 17, 2010
|
|
4K5C
 
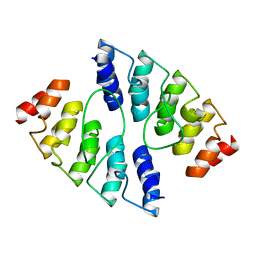 | |
4K5B
 
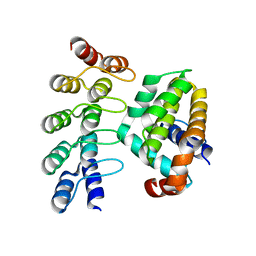 | | Co-crystallization with conformation-specific designed ankyrin repeat proteins explains the conformational flexibility of BCL-W | | Descriptor: | Apoptosis regulator BCL-W, Bcl-2-like protein 2 | | Authors: | Schilling, J, Schoeppe, J, Sauer, E, Plueckthun, A. | | Deposit date: | 2013-04-14 | | Release date: | 2014-04-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Co-Crystallization with Conformation-Specific Designed Ankyrin Repeat Proteins Explains the Conformational Flexibility of BCL-W
J.Mol.Biol., 426, 2014
|
|
2YEQ
 
 | | Structure of PhoD | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ALKALINE PHOSPHATASE D, ... | | Authors: | Lillington, J.E.D, Rodriguez, F, Roversi, P, Johnson, S.J, Berks, B, Lea, S.M. | | Deposit date: | 2011-03-30 | | Release date: | 2012-04-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure of the Bacillus Subtilis Phosphodiesterase Phod Reveals an Iron and Calcium-Containing Active Site.
J.Biol.Chem., 289, 2014
|
|
6BBN
 
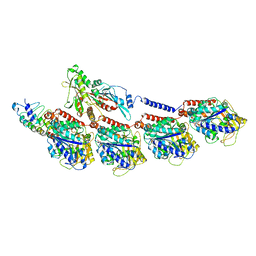 | |
3Q8J
 
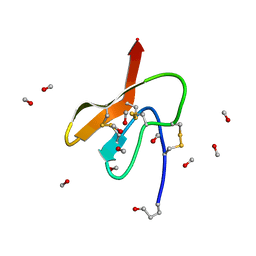 | | Crystal Structure of Asteropsin A from Marine Sponge Asteropus sp. | | Descriptor: | Asteropsin A, METHANOL | | Authors: | Bowling, J.J, Fronczek, F.R, Hamann, M.T, Li, H, Jung, J.H. | | Deposit date: | 2011-01-06 | | Release date: | 2012-04-18 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (0.87 Å) | | Cite: | An Uncommon Crystal Structure of a Marine Knottin Peptide from Asteropus sp.
To be published
|
|
3BZ9
 
 | | Crystal Structures of (S)-(-)-Blebbistatin Analogs bound to Dictyostelium discoideum myosin II | | Descriptor: | (3aS)-3a-hydroxy-1-phenyl-1,2,3,3a-tetrahydro-4H-pyrrolo[2,3-b]quinolin-4-one, 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Allingham, J.S, Rayment, I. | | Deposit date: | 2008-01-17 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The small molecule tool (S)-(-)-blebbistatin: novel insights of relevance to myosin inhibitor design.
Org.Biomol.Chem., 6, 2008
|
|
2XGA
 
 | | MTSL spin-labelled Shigella Flexneri Spa15 | | Descriptor: | S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, SURFACE PRESENTATION OF ANTIGENS PROTEIN SPAK | | Authors: | Lillington, J.E.D, Johnson, S, Lea, S.M. | | Deposit date: | 2010-06-02 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Shigella Flexneri Spa15 Crystal Structure Verified in Solution by Double Electron Electron Resonance.
J.Mol.Biol., 405, 2011
|
|
3BZ8
 
 | | Crystal Structures of (S)-(-)-Blebbistatin Analogs bound to Dictyostelium discoideum myosin II | | Descriptor: | (3aS)-3a-hydroxy-7-methyl-1-phenyl-1,2,3,3a-tetrahydro-4H-pyrrolo[2,3-b]quinolin-4-one, 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Allingham, J.S, Rayment, I. | | Deposit date: | 2008-01-17 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The small molecule tool (S)-(-)-blebbistatin: novel insights of relevance to myosin inhibitor design.
Org.Biomol.Chem., 6, 2008
|
|
