4N7R
 
 | | Crystal structure of Arabidopsis glutamyl-tRNA reductase in complex with its binding protein | | Descriptor: | Genomic DNA, chromosome 3, P1 clone: MXL8, ... | | Authors: | Zhao, A, Fang, Y, Lin, Y, Gong, W, Liu, L. | | Deposit date: | 2013-10-16 | | Release date: | 2014-05-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Crystal structure of Arabidopsis glutamyl-tRNA reductase in complex with its stimulator protein
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3EFZ
 
 | | Crystal Structure of a 14-3-3 protein from cryptosporidium parvum (cgd1_2980) | | Descriptor: | 1,2-ETHANEDIOL, 14-3-3 protein | | Authors: | Wernimont, A.K, Dong, A, Qiu, W, Lew, J, Wasney, G.A, Vedadi, M, Kozieradzki, I, Zhao, Y, Ren, H, Alam, Z, Lin, Y.H, Sundstrom, M, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Hui, R, Brokx, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-10 | | Release date: | 2008-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Characterization of 14-3-3 proteins from Cryptosporidium parvum.
Plos One, 6, 2011
|
|
4NAQ
 
 | | Crystal structure of porcine aminopeptidase-N complexed with poly-alanine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Chen, L, Lin, Y.L, Peng, G, Li, F. | | Deposit date: | 2013-10-22 | | Release date: | 2013-12-04 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for multifunctional roles of mammalian aminopeptidase N.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5U6I
 
 | | Discovery of MLi-2, an Orally Available and Selective LRRK2 Inhibitor that Reduces Brain Kinase Activity | | Descriptor: | 3-[2-(morpholin-4-yl)pyridin-4-yl]-5-[(propan-2-yl)oxy]-1H-indazole, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Scott, J.D, DeMong, D.E, Fell, M.J, Mirescu, C, Basu, K, Greshock, T.J, Morrow, J.A, Xiao, L, Hruza, A, Harris, J, Tiscia, H.E, Chang, R.K, Embrey, M.W, McCauley, J.A, Li, W, Lin, S, Liu, H, Dai, X, Baptista, M, Agnihotri, G, Columbus, J, Mei, H, Poirier, M, Zhou, X, Lin, Y, Yin, Z, Sanders, J.M, Drolet, R.E, Kern, J.T, Kennedy, M.E, Parker, E.M, Stamford, A.W, Nargund, R, Miller, M.W. | | Deposit date: | 2016-12-08 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of a 3-(4-Pyrimidinyl) Indazole (MLi-2), an Orally Available and Selective Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitor that Reduces Brain Kinase Activity.
J. Med. Chem., 60, 2017
|
|
4B3Z
 
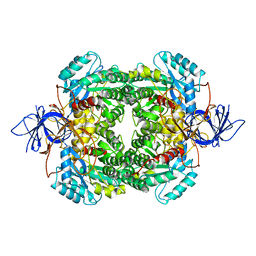 | | Structure of the human collapsin response mediator protein-1, a lung cancer suppressor | | Descriptor: | DIHYDROPYRIMIDINASE-RELATED PROTEIN 1 | | Authors: | Liu, S.H, Lin, Y.H, Huang, S.F, Niou, Y.K, Huang, L.L, Chen, Y.J. | | Deposit date: | 2012-07-27 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure of Human Collapsin Response Mediator Protein 1: A Possible Role of its C-Terminal Tail.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
2AZC
 
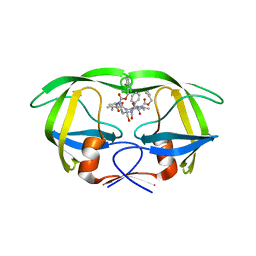 | | HIV-1 Protease NL4-3 6X mutant | | Descriptor: | PROTEASE RETROPEPSIN, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Heaslet, H, Kutilek, V, Morris, G.M, Lin, Y.-C, Elder, J.H, Torbett, B.E, Stout, C.D. | | Deposit date: | 2005-09-10 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Insights into the Mechanisms of Drug Resistance in HIV-1 Protease NL4-3
J.Mol.Biol., 356, 2006
|
|
4NZ8
 
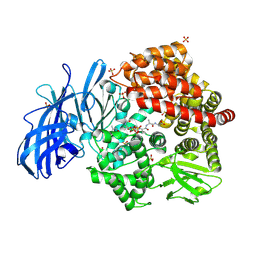 | | Crystal structure of porcine aminopeptidase-N complexed with cleaved poly-alanine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Chen, L, Lin, Y.L, Peng, G, Li, F. | | Deposit date: | 2013-12-11 | | Release date: | 2013-12-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for multifunctional roles of mammalian aminopeptidase N.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2BA3
 
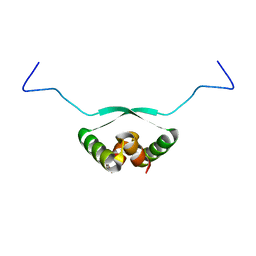 | | NMR Structure of NikA N-terminal Fragment | | Descriptor: | NikA | | Authors: | Yoshida, H, Furuya, N, Lin, Y.J, Guntert, P, Komano, T, Kainosho, M. | | Deposit date: | 2005-10-14 | | Release date: | 2006-10-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of NikA N-teminal Fragment
To be Published
|
|
3EB0
 
 | | Crystal Structure of cgd4_240 from cryptosporidium Parvum in complex with indirubin E804 | | Descriptor: | 3-({[(3S)-3,4-dihydroxybutyl]oxy}amino)-1H,2'H-2,3'-biindol-2'-one, GLYCEROL, Putative uncharacterized protein | | Authors: | Wernimont, A.K, Fedorov, O, Lam, A, Ali, A, Zhao, Y, Lew, J, Wasney, G, Vedadi, M, Kozieradzki, I, Schapira, M, Bochkarev, A, Wilstrom, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-08-26 | | Release date: | 2008-09-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of cgd4_240 from cryptosporidium Parvum in complex with indirubin E804
TO BE PUBLISHED
|
|
3Q5L
 
 | | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K 213 in the presence of 17-AEP-geldanamycin | | Descriptor: | (4E,6Z,8S,9S,10E,12S,13R,14S,16R)-13-hydroxy-8,14-dimethoxy-4,10,12,16-tetramethyl-3,20,22-trioxo-19-{[2-(pyrrolidin-1-yl)ethyl]amino}-2-azabicyclo[16.3.1]docosa-1(21),4,6,10,18-pentaen-9-yl carbamate, Heat shock protein 83-1 | | Authors: | Wernimont, A.K, Tempel, W, Lin, Y.H, Hutchinson, A, MacKenzie, F, Fairlamb, A, Cossar, D, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-28 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K 213 in the presence of 17-AEP-geldanamycin.
To be Published
|
|
3E5R
 
 | |
3E95
 
 | | Crystal Structure of the Plasmodium Falciparum ubiquitin conjugating enzyme complex, PfUBC13-PfUev1a | | Descriptor: | UNKNOWN ATOM OR ION, Ubiquitin carrier protein, Ubiquitin-conjugating enzyme E2 | | Authors: | Wernimont, A.K, Lam, A, Ali, A, Brokx, S, Lin, Y.H, Zhao, Y, Lew, J, Ravichandran, M, Wasney, G, Vedadi, M, Kozieradzki, I, Schapira, M, Bochkarev, A, Wilkstrom, M, BOuntra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Qiu, W, Brand, V.B, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-08-21 | | Release date: | 2008-09-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Plasmodium Falciparum ubiquitin conjugating enzyme complex, PfUBC13-PfUev1a
TO BE PUBLISHED
|
|
3E6A
 
 | |
7EW8
 
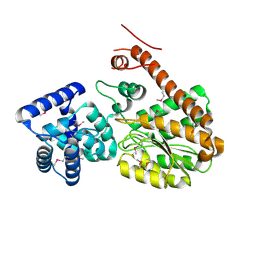 | | Legionella pneumophila effector AnkD | | Descriptor: | ANK_REP_REGION domain-containing protein | | Authors: | Chen, T.T, Lin, Y.L. | | Deposit date: | 2021-05-24 | | Release date: | 2022-06-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Atypical Legionella GTPase effector hijacks host vesicular transport factor p115 to regulate host lipid droplet.
Sci Adv, 8, 2022
|
|
3SHB
 
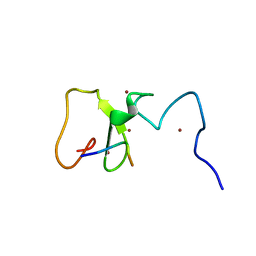 | | Crystal Structure of PHD Domain of UHRF1 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Histone H3 peptide, ZINC ION | | Authors: | Hu, L, Li, Z, Wang, P, Lin, Y, Xu, Y. | | Deposit date: | 2011-06-16 | | Release date: | 2011-08-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of PHD domain of UHRF1 and insights into recognition of unmodified histone H3 arginine residue 2.
Cell Res., 2011
|
|
3F9R
 
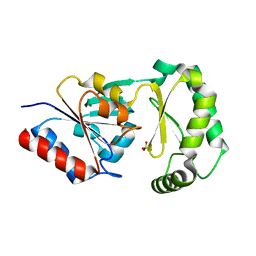 | | Crystal Structure of Trypanosoma Brucei phosphomannosemutase, TB.10.700.370 | | Descriptor: | MAGNESIUM ION, Phosphomannomutase, SULFATE ION | | Authors: | Wernimont, A.K, Lam, A, Ali, A, Lin, Y.H, Guther, L, Shamshad, A, MacKenzie, F, Bandini, G, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-14 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Trypanosoma Brucei phosphomannosemutase, TB.10.700.370
To be Published
|
|
7WX6
 
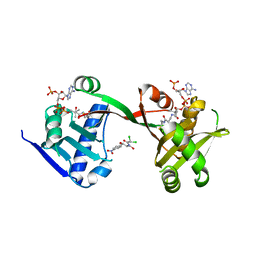 | | A Legionella acetyltransferase VipF | | Descriptor: | CHLORAMPHENICOL, COENZYME A, N-acetyltransferase | | Authors: | Chen, T.T, Lin, Y.L, Chen, Z, Han, A.D. | | Deposit date: | 2022-02-14 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.273 Å) | | Cite: | Structural basis for the acetylation mechanism of the Legionella effector VipF.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7WX7
 
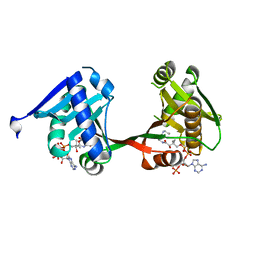 | | complex of a legionella acetyltransferase VipF and COA/ACO | | Descriptor: | ACETYL COENZYME *A, COENZYME A, N-acetyltransferase | | Authors: | Chen, T.T, Lin, Y.L, Zhang, S.J, Han, A.D. | | Deposit date: | 2022-02-14 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | Structural basis for the acetylation mechanism of the Legionella effector VipF.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7WX5
 
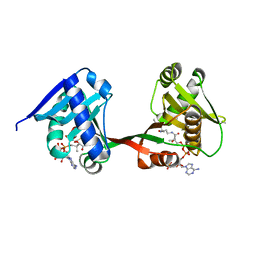 | | a Legionella acetyltransferase effector VipF | | Descriptor: | ACETYL COENZYME *A, N-acetyltransferase | | Authors: | Chen, T.T, Lin, Y.L, Zhang, S.J, Han, A.D. | | Deposit date: | 2022-02-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Structural basis for the acetylation mechanism of the Legionella effector VipF.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
5C8G
 
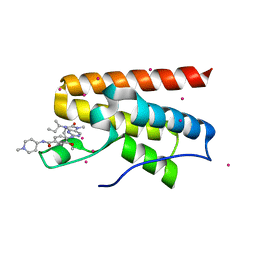 | | Crystal Structure Analysis of PP-BRD20 from Tb427tmp complexed with BI-2536 | | Descriptor: | 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, PP-BRD20, UNKNOWN ATOM OR ION | | Authors: | Jiang, D.Q, Tempel, W, Loppnau, P, Graslund, S, Hou, C.F.D, El Bakkouri, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Amain, M, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-25 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure Analysis of PP-BRD20 from Tb427tmp
to be published
|
|
3GUE
 
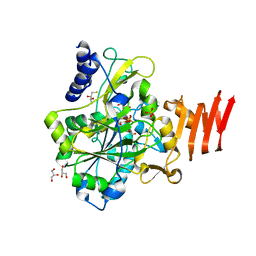 | | Crystal Structure of UDP-glucose phosphorylase from Trypanosoma Brucei, (Tb10.389.0330) | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Wernimont, A.K, Marino, K, Lin, Y.H, Mackenzie, F, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-03-29 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of UDP-glucose phosphorylase from Trypanosoma Brucei, (Tb10.389.0330)
To be Published
|
|
3H80
 
 | | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LmjF33.0312:M1-K213 | | Descriptor: | 1,2-ETHANEDIOL, Heat shock protein 83-1, MAGNESIUM ION, ... | | Authors: | Wernimont, A.K, Tempel, W, Lin, Y.H, Hutchinson, A, Mackenzie, F, Fairlamb, A, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-28 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LmjF33.0312:M1-K213
To be Published
|
|
5CHZ
 
 | |
7XQL
 
 | | complex structure of LegA15 with GNP | | Descriptor: | Ankyrin repeat-containing protein, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Chen, T.T, Lin, Y.L. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.272 Å) | | Cite: | Atypical Legionella GTPase effector hijacks host vesicular transport factor p115 to regulate host lipid droplet.
Sci Adv, 8, 2022
|
|
3FB3
 
 | | Crystal Structure of Trypanosoma Brucei Acetyltransferase, Tb11.01.2886 | | Descriptor: | N-acetyltransferase | | Authors: | Wernimont, A.K, Marino, K, Zhang, A.Z, Ma, D, Lin, Y.H, MacKenzie, F, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-18 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Trypanosoma Brucei Acetyltransferase, Tb11.01.2886
TO BE PUBLISHED
|
|
