4IFN
 
 | | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes | | Descriptor: | (1R,2R)-2-{[(1S)-1-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)methyl]-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}cyclohexanecarboxylic acid, kelch-like ECH-associated protein 1 | | Authors: | Pan, H, Lin, M, Yang, Y, Callaway, K, Baker, J, Diep, L, Yan, J, Tanaka, K, Zhu, Y.L, Konradi, A.W, Jobling, M, Tam, D, Ren, Z, Cheung, H, Bova, M, Riley, B.E, Yao, N, Artis, D.R. | | Deposit date: | 2012-12-14 | | Release date: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes
Acta Crystallogr.,Sect.D
|
|
6Z0F
 
 | | Crystal structure of the membrane pseudokinase YukC/EssB from Bacillus subtilis T7SS | | Descriptor: | ESX secretion system protein YukC | | Authors: | Tassinari, M, Bellinzoni, M, Alzari, P.M, Fronzes, R, Gubellini, F. | | Deposit date: | 2020-05-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | The Antibacterial Type VII Secretion System of Bacillus subtilis: Structure and Interactions of the Pseudokinase YukC/EssB.
Mbio, 13, 2022
|
|
4IFJ
 
 | | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes | | Descriptor: | Kelch-like ECH-associated protein 1 | | Authors: | Pan, H, Lin, M, Yang, Y, Callaway, K, Baker, J, Diep, L, Yan, J, Tanaka, K, Zhu, Y.L, Konradi, A.W, Jobling, M, Tam, D, Ren, Z, Cheung, H, Bova, M, Riley, B.E, Yao, N, Artis, D.R. | | Deposit date: | 2012-12-14 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes
To be Published
|
|
4IFL
 
 | | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes | | Descriptor: | Nrf2 peptide, kelch-like ECH-associated protein 1 | | Authors: | Pan, H, Lin, M, Yang, Y, Callaway, K, Baker, J, Diep, L, Yan, J, Tanaka, K, Zhu, Y.L, Konradi, A.W, Jobling, M, Tam, D, Ren, Z, Cheung, H, Bova, M, Riley, B.E, Yao, N, Artis, D.R. | | Deposit date: | 2012-12-14 | | Release date: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes
Acta Crystallogr.,Sect.D
|
|
6WEM
 
 | | Crimson 0.9 | | Descriptor: | mCrimson 0.9 | | Authors: | Ataie, N, Tran Tang, C, Sens, A, Lin, M.Z, Chu, J, Ng, H.L. | | Deposit date: | 2020-04-02 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crimson 0.9
To Be Published
|
|
2ABI
 
 | | Crystal structure of the human mineralocorticoid receptor ligand-binding domain bound to deoxycorticosterone | | Descriptor: | DESOXYCORTICOSTERONE, Mineralocorticoid receptor | | Authors: | Huyet, J, Pinon, G.-M, Rochel, M, Mayer, C, Rafestin-Oblin, M.-E, Fagart, J. | | Deposit date: | 2005-07-15 | | Release date: | 2006-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of the human mineralocorticoid receptor ligand-binding domain bound to deoxycorticosterone
To be published
|
|
8H3G
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) E166V Mutant in Complex with Inhibitor Enstrelvir | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, GLYCEROL | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H3L
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (T21I and E166V) in Complex with Inhibitor Enstrelvir | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H3K
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (L50F and E166V) in Complex with Inhibitor Enstrelvir | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, ... | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
5YBA
 
 | | Dimeric Cyclophilin from T.vaginalis in complex with Myb1 peptide | | Descriptor: | Myb1 peptide, Peptidyl-prolyl cis-trans isomerase | | Authors: | Cho, C.C, Lin, M.H, Martin, T, Chou, C.C, Chen, C, Hsu, C.H. | | Deposit date: | 2017-09-04 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.062 Å) | | Cite: | Structural basis of interaction between dimeric cyclophilin 1 and Myb1 transcription factor in Trichomonas vaginalis
Sci Rep, 8, 2018
|
|
5BQL
 
 | | Fluorescent protein cyOFP | | Descriptor: | Fluorescent protein cyOFP | | Authors: | Sens, A, Ataie, N, Ng, H.L, Lin, M.Z, Chu, J. | | Deposit date: | 2015-05-29 | | Release date: | 2016-06-01 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | A Guide to Fluorescent Protein FRET Pairs.
Sensors Basel Sensors, 16, 2016
|
|
8I0J
 
 | | JB13GH39P28 mutant-D41G | | Descriptor: | CHLORIDE ION, Glycoside hydrolase family 39 beta-xylosidase | | Authors: | Zhou, J.P, Cao, L.J, Lin, M.Y, Zhang, R, Huang, Z.X. | | Deposit date: | 2023-01-11 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | beta-Xylosidase JB13GH39P28 (D41G) showing salt/ethanol/trypsin tolerance and transformation of notoginsenosides
To Be Published
|
|
8I0X
 
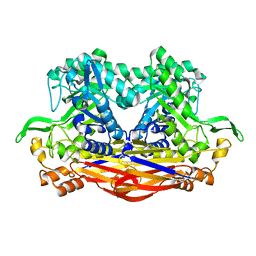 | | Beta-Xylosidase JB13GH39P28 showing salt/ethanol/trypsin tolerance, low-pH/low-Temperature activity, and transformation of notoginsenosides | | Descriptor: | Glycoside hydrolase family 39 beta-xylosidase | | Authors: | Zhou, J.P, Cao, L.J, Lin, M.Y, Zhang, R, Huang, Z.X. | | Deposit date: | 2023-01-11 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | beta-Xylosidase JB13GH39P28(D41G)showing salt/ethanol/trypsin tolerance and transformation of notoginsenosides
To Be Published
|
|
5YB9
 
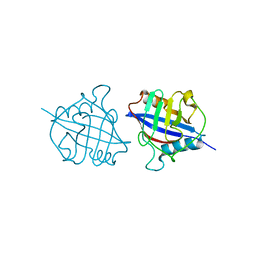 | | Crystal structure of a dimeric cyclophilin A from T.vaginalis | | Descriptor: | Peptidyl-prolyl cis-trans isomerase | | Authors: | Cho, C.C, Lin, M.H, Chou, C.C, Martin, T, Chen, C, Hsu, C.H. | | Deposit date: | 2017-09-04 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.276 Å) | | Cite: | Structural basis of interaction between dimeric cyclophilin 1 and Myb1 transcription factor in Trichomonas vaginalis
Sci Rep, 8, 2018
|
|
3B6E
 
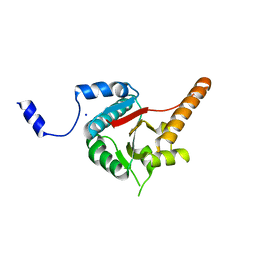 | | Crystal structure of human DECH-box RNA Helicase MDA5 (Melanoma differentiation-associated protein 5), DECH-domain | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, SODIUM ION | | Authors: | Karlberg, T, Welin, M, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Human DECH-box RNA Helicase MDA5 (Melanoma differentiation-associated protein 5), DECH-domain.
To be Published
|
|
3B7K
 
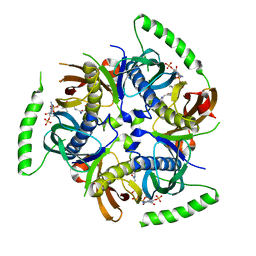 | | Human Acyl-coenzyme A thioesterase 12 | | Descriptor: | Acyl-coenzyme A thioesterase 12, COENZYME A | | Authors: | Lehtio, L, Busam, R.D, Arrowsmith, C.H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Berglund, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-31 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human Acyl-coenzyme A thioesterase 12.
To be Published
|
|
5JKI
 
 | | Crystal structure of the first transmembrane PAP2 type phosphatidylglycerolphosphate phosphatase from Bacillus subtilis | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Putative lipid phosphate phosphatase YodM, TUNGSTATE(VI)ION, ... | | Authors: | El Ghachi, M, Howe, N, Lampion, A, Delbrassine, F, Vogeley, L, Caffrey, M, Sauvage, E, Auger, R, Guiseppe, A, Roure, S, Perlier, S, Mengin-lecreulx, D, Foglino, M, Touze, T. | | Deposit date: | 2016-04-26 | | Release date: | 2017-02-22 | | Last modified: | 2017-05-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure and biochemical characterization of the transmembrane PAP2 type phosphatidylglycerol phosphate phosphatase from Bacillus subtilis.
Cell. Mol. Life Sci., 74, 2017
|
|
5DMA
 
 | |
3BIN
 
 | | Structure of the DAL-1 and TSLC1 (372-383) complex | | Descriptor: | Band 4.1-like protein 3, Cell adhesion molecule 1 | | Authors: | Busam, R.D, Arrowsmith, C.H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Berglund, H, Persson, C, Hallberg, B.M. | | Deposit date: | 2007-11-30 | | Release date: | 2008-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of tumor suppressor in lung cancer 1 (TSLC1) binding to differentially expressed in adenocarcinoma of the lung (DAL-1/4.1B)
J.Biol.Chem., 286, 2011
|
|
4WB6
 
 | | Crystal structure of a L205R mutant of human cAMP-dependent protein kinase A (catalytic alpha subunit) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PKI (5-24), ... | | Authors: | Cheung, J, Ginter, C, Cassidy, M, Franklin, M.C, Rudolph, M.J, Hendrickson, W.A. | | Deposit date: | 2014-09-02 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into mis-regulation of protein kinase A in human tumors.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3CE0
 
 | | Human poly(ADP-ribose) polymerase 3, catalytic fragment in complex with an inhibitor PJ34 | | Descriptor: | N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE, Poly [ADP-ribose] polymerase 3 | | Authors: | Lehtio, L, Karlberg, T, Arrowsmith, C.H, Berglund, H, Bountra, C, Busam, R, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, van den Berg, S, Welin, M, Weigelt, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-27 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for inhibitor specificity in human poly(ADP-ribose) polymerase-3.
J.Med.Chem., 52, 2009
|
|
3BLJ
 
 | | Crystal structure of human poly(ADP-ribose) polymerase 15, catalytic fragment | | Descriptor: | CHLORIDE ION, GLYCEROL, Poly(ADP-ribose) polymerase 15, ... | | Authors: | Karlberg, T, Lehtio, L, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Welin, M, Weigelt, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-12-11 | | Release date: | 2007-12-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Lack of ADP-ribosyltransferase Activity in Poly(ADP-ribose) Polymerase-13/Zinc Finger Antiviral Protein.
J.Biol.Chem., 290, 2015
|
|
6D39
 
 | |
6D38
 
 | |
5DUS
 
 | | Crystal structure of MERS-CoV macro domain in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, GLYCEROL, ORF1a, ... | | Authors: | Cho, C.-C, Lin, M.-H, Chuang, C.-Y, Hsu, C.-H. | | Deposit date: | 2015-09-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.432 Å) | | Cite: | Macro Domain from Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Is an Efficient ADP-ribose Binding Module: CRYSTAL STRUCTURE AND BIOCHEMICAL STUDIES
J.Biol.Chem., 291, 2016
|
|
