6KIM
 
 | |
7YKF
 
 | | Crystal structure of MAGI2 PDZ0-GK/pEphexin4 complex | | Descriptor: | Ephexin4, Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 | | Authors: | Zhang, M, Lin, L, Zhu, J. | | Deposit date: | 2022-07-22 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Phosphorylation-dependent recognition of diverse protein targets by the cryptic GK domain of MAGI MAGUKs.
Sci Adv, 9, 2023
|
|
7YKI
 
 | | Crystal structure of MAGI2 PDZ0-GK domain in complex with phospho-SAPAP1 GBR3 peptide | | Descriptor: | GLYCEROL, Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2, ... | | Authors: | Zhang, M, Lin, L, Zhu, J. | | Deposit date: | 2022-07-22 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphorylation-dependent recognition of diverse protein targets by the cryptic GK domain of MAGI MAGUKs.
Sci Adv, 9, 2023
|
|
7YKG
 
 | | Crystal structure of MAGI2 PDZ0-GK/pSGEF complex | | Descriptor: | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2, SGEF | | Authors: | Zhang, M, Lin, L, Zhu, J. | | Deposit date: | 2022-07-22 | | Release date: | 2023-08-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Phosphorylation-dependent recognition of diverse protein targets by the cryptic GK domain of MAGI MAGUKs.
Sci Adv, 9, 2023
|
|
7YKH
 
 | | Crystal structure of MAGI2 PDZ0-GK domain in complex with phospho-SAPAP1 GBR2 peptide | | Descriptor: | GLYCEROL, Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2, ... | | Authors: | Zhang, M, Lin, L, Zhu, J. | | Deposit date: | 2022-07-22 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Phosphorylation-dependent recognition of diverse protein targets by the cryptic GK domain of MAGI MAGUKs.
Sci Adv, 9, 2023
|
|
4XSK
 
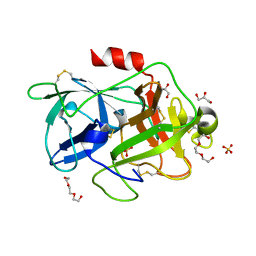 | | Structure of PAItrap, an uPA mutant | | Descriptor: | GLYCEROL, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Gong, L, Proulle, V, Hong, Z, Lin, Z, Liu, M, Yuan, C, Lin, L, Furie, B, Flaumenhaft, R, Andreasen, P, Furie, B, Huang, M. | | Deposit date: | 2015-01-22 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of PAItrap, an uPA mutant
To Be Published
|
|
6Z4Q
 
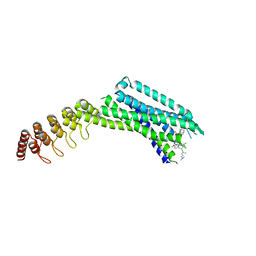 | | Crystal structure of the neurotensin receptor 1 in complex with the small-molecule inverse agonist SR142948A | | Descriptor: | 2-[[5-(2,6-dimethoxyphenyl)-1-[4-[3-(dimethylamino)propyl-methyl-carbamoyl]-2-propan-2-yl-phenyl]pyrazol-3-yl]carbonylamino]adamantane-2-carboxylic acid, Neurotensin receptor type 1,Neurotensin receptor type 1,DARPin | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-05-25 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.923 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6ZA8
 
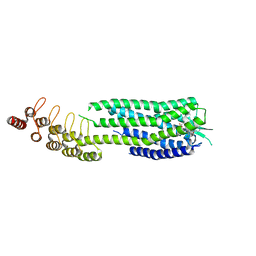 | | Crystal structure of the neurotensin receptor 1 in complex with the small-molecule partial agonist RTI-3a | | Descriptor: | (2~{S})-2-[[1-(7-chloranylquinolin-4-yl)-5-(2,6-dimethoxyphenyl)pyrazol-3-yl]carbonylamino]-4-methyl-pentanoic acid, Neurotensin receptor type 1,Neurotensin receptor type 1,Neurotensin receptor 1 (NTSR1),Neurotensin receptor 1 (NTSR1),Neurotensin receptor type 1,Neurotensin receptor 1 (NTSR1),Neurotensin receptor 1 (NTSR1) | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-06-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6ZIN
 
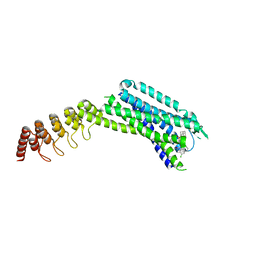 | | Crystal structure of the neurotensin receptor 1 in complex with the small molecule inverse agonist SR48692 | | Descriptor: | 2-[[1-(7-chloranylquinolin-4-yl)-5-(2,6-dimethoxyphenyl)pyrazol-3-yl]carbonylamino]adamantane-2-carboxylic acid, Neurotensin receptor type 1,DARPin,HRV 3C protease recognition sequence | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-06-26 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.639 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6Z8N
 
 | | Crystal structure of the neurotensin receptor 1 in complex with the small-molecule full agonist SRI-9829 | | Descriptor: | (2~{S})-4-methyl-2-[(1-quinolin-8-ylsulfonylindol-3-yl)carbonylamino]pentanoic acid, Neurotensin receptor type 1,Neurotensin receptor type 1,Neurotensin receptor type 1,Neurotensin receptor type 1,Neurotensin receptor 1 (NTSR1),Neurotensin receptor 1 (NTSR1) | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-06-02 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6Z4V
 
 | | Crystal structure of the neurotensin receptor 1 (NTSR1-H4bmx) in complex with NTS8-13 | | Descriptor: | ARG-PRO-TYR-ILE-LEU, Neurotensin receptor type 1,Neurotensin receptor type 1,DARPin | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-05-25 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6YVR
 
 | | Crystal structure of the neurotensin receptor 1 in complex with the peptide full agonist NTS8-13 | | Descriptor: | Neurotensin receptor type 1,Neurotensin receptor type 1,DARPin crystallisation chaperone, neurotensin NTS8-13 (full agonist), nonyl beta-D-glucopyranoside | | Authors: | Deluigi, M, Merklinger, L, Hilge, M, Ernst, P, Klipp, A, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-04-28 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.458 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6Z4S
 
 | | Crystal structure of the neurotensin receptor 1 (NTSR1-H4bmx) in complex with the small molecule inverse agonist SR48692 | | Descriptor: | 2-[[1-(7-chloranylquinolin-4-yl)-5-(2,6-dimethoxyphenyl)pyrazol-3-yl]carbonylamino]adamantane-2-carboxylic acid, Neurotensin receptor type 1,DARPin,HRV 3C protease recognition sequence | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-05-25 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.707 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6Z66
 
 | | Crystal structure of apo-state neurotensin receptor 1 | | Descriptor: | Neurotensin receptor type 1,Neurotensin receptor type 1,DARPin | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-05-27 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.192 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
4DHS
 
 | | Small-molecule inhibitors of 14-3-3 protein-protein interactions from virtual screening | | Descriptor: | (2-{2-[(3,5-dichlorophenyl)amino]-2-oxoethoxy}phenyl)phosphonic acid, 14-3-3 PROTEIN SIGMA, CHLORIDE ION, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2012-01-30 | | Release date: | 2013-07-31 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions.
Chem.Commun.(Camb.), 49, 2013
|
|
4DHN
 
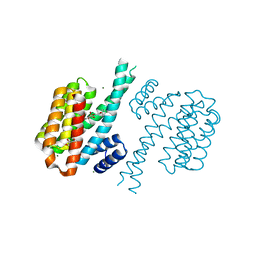 | | Small-molecule inhibitors of 14-3-3 protein-protein interactions from virtual screening | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2012-01-30 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions.
Chem.Commun.(Camb.), 49, 2013
|
|
5Z9J
 
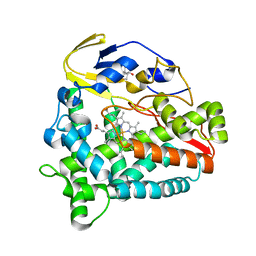 | | Identification of the functions of unusual cytochrome p450-like monooxygenases involved in microbial secondary metablism | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative P450-like enzyme, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Lu, M, Lin, L, Zhang, C, Chen, Y. | | Deposit date: | 2018-02-03 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Riboflavin Is Directly Involved in the N-Dealkylation Catalyzed by Bacterial Cytochrome P450 Monooxygenases.
Chembiochem, 2020
|
|
5ZXL
 
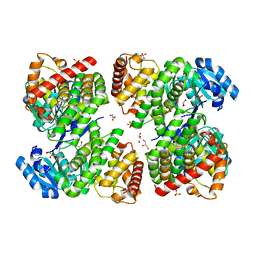 | | Structure of GldA from E.coli | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycerol dehydrogenase, ... | | Authors: | Zhang, J, Lin, L. | | Deposit date: | 2018-05-21 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Structure of glycerol dehydrogenase (GldA) from Escherichia coli.
Acta Crystallogr F Struct Biol Commun, 75, 2019
|
|
5Z9I
 
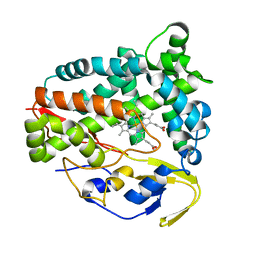 | |
3NEN
 
 | |
4MAD
 
 | | Crystal structure of beta-galactosidase C (BgaC) from Bacillus circulans ATCC 31382 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Beta-galactosidase | | Authors: | Kamerke, C, You, D.J, Kanaya, S, Elling, L. | | Deposit date: | 2013-08-16 | | Release date: | 2014-08-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational design of a glycosynthase by the crystal structure of beta-galactosidase from Bacillus circulans (BgaC) and its use for the synthesis of N-acetyllactosamine type 1 glycan structures.
J.Biotechnol., 191, 2014
|
|
1F7L
 
 | | HOLO-(ACYL CARRIER PROTEIN) SYNTHASE IN COMPLEX WITH COENZYME A AT 1.5A | | Descriptor: | CALCIUM ION, CHLORIDE ION, COENZYME A, ... | | Authors: | Parris, K.D, Lin, L, Tam, A, Mathew, R, Hixon, J, Stahl, M, Fritz, C.C, Seehra, J, Somers, W.S. | | Deposit date: | 2000-06-27 | | Release date: | 2001-06-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of substrate binding to Bacillus subtilis holo-(acyl carrier protein) synthase reveal a novel trimeric arrangement of molecules resulting in three active sites.
Structure Fold.Des., 8, 2000
|
|
1F7T
 
 | | HOLO-(ACYL CARRIER PROTEIN) SYNTHASE AT 1.8A | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Parris, K.D, Lin, L, Tam, A, Mathew, R, Hixon, J, Stahl, M, Fritz, C.C, Seehra, J, Somers, W.S. | | Deposit date: | 2000-06-27 | | Release date: | 2001-06-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of substrate binding to Bacillus subtilis holo-(acyl carrier protein) synthase reveal a novel trimeric arrangement of molecules resulting in three active sites.
Structure Fold.Des., 8, 2000
|
|
1F80
 
 | | HOLO-(ACYL CARRIER PROTEIN) SYNTHASE IN COMPLEX WITH HOLO-(ACYL CARRIER PROTEIN) | | Descriptor: | ACYL CARRIER PROTEIN, HOLO-(ACYL CARRIER PROTEIN) SYNTHASE, SODIUM ION | | Authors: | Parris, K.D, Lin, L, Tam, A, Mathew, R, Hixon, J, Stahl, M, Fritz, C.C, Seehra, J, Somers, W.S. | | Deposit date: | 2000-06-28 | | Release date: | 2001-06-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of substrate binding to Bacillus subtilis holo-(acyl carrier protein) synthase reveal a novel trimeric arrangement of molecules resulting in three active sites.
Structure Fold.Des., 8, 2000
|
|
7E12
 
 | | Crystal structure of PKAc-A11E complex | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, THR-ARG-SER-GLU-ILE-ARG-ARG-ALA-SER-THR-ILE-GLU, ... | | Authors: | Qin, J, Lin, L, Yuchi, Z. | | Deposit date: | 2021-01-28 | | Release date: | 2022-04-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Structures of PKA-phospholamban complexes reveal a mechanism of familial dilated cardiomyopathy.
Elife, 11, 2022
|
|
