4GVT
 
 | | Crystal structure of D48V mutant of human GLTP bound with 12:0 disulfatide (hexagonal form) | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-1-[(3,6-di-O-sulfo-beta-D-galactopyranosyl)oxy]-3-hydroxyoctadec-4-en-2-yl}dodecanamide | | Authors: | Samygina, V.R, Cabo-Bilbao, A, Goni-de-Cerio, F, Popov, A.N, Malinina, L. | | Deposit date: | 2012-08-31 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GSO
 
 | | structure of Jararacussin-I | | Descriptor: | Thrombin-like enzyme BjussuSP-1 | | Authors: | Ullah, A, Souza, T.C.A.B, Zanphorlin, L.M, Mariutti, R, Sanata, S.V, Murakami, M.T, Arni, R.K. | | Deposit date: | 2012-08-28 | | Release date: | 2012-12-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Jararacussin-I: The highly negatively charged catalytic interface contributes to macromolecular selectivity in snake venom thrombin-like enzymes.
Protein Sci., 22, 2013
|
|
1NGT
 
 | | The Role of Minor Groove Functional Groups in DNA Hydration | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*(MTR)P*TP*CP*GP*CP*G)-3', MAGNESIUM ION | | Authors: | Woods, K.K, Lan, T, McLaughlin, L.W, Williams, L.D. | | Deposit date: | 2002-12-17 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The Role of Minor Groove Functional Groups in DNA Hydration
Nucleic Acids Res., 31, 2003
|
|
4E48
 
 | |
1BY5
 
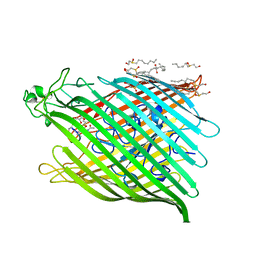 | | FHUA FROM E. COLI, WITH ITS LIGAND FERRICHROME | | Descriptor: | FE (III) ION, FERRIC HYDROXAMATE UPTAKE PROTEIN, FERRICHROME, ... | | Authors: | Locher, K.P, Rees, B, Koebnik, R, Mitschler, A, Moulinier, L, Rosenbusch, J.P, Moras, D. | | Deposit date: | 1998-10-23 | | Release date: | 1999-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes.
Cell(Cambridge,Mass.), 95, 1998
|
|
465D
 
 | | STRUCTURE OF THE TOPOISOMERASE II POISON BOUND TO DNA | | Descriptor: | 9-AMINO-(N-(2-DIMETHYLAMINO)ETHYL)ACRIDINE-4-CARBOXAMIDE, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3') | | Authors: | Adams, A, Guss, J.M, Collyer, C.A, Denny, W.A, Wakelin, L.P. | | Deposit date: | 1999-04-14 | | Release date: | 1999-08-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the topoisomerase II poison 9-amino-[N-(2-dimethylamino)ethyl]acridine-4-carboxamide bound to the DNA hexanucleotide d(CGTACG)2.
Biochemistry, 38, 1999
|
|
4GIX
 
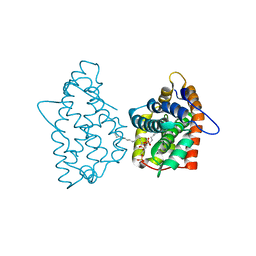 | | Crystal structure of human GLTP bound with 12:0 disulfatide | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-1-[(3,6-di-O-sulfo-beta-D-galactopyranosyl)oxy]-3-hydroxyoctadec-4-en-2-yl}dodecanamide | | Authors: | Samygina, V.R, Ochoa-Lizarralde, B, Malinina, L. | | Deposit date: | 2012-08-09 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3RIC
 
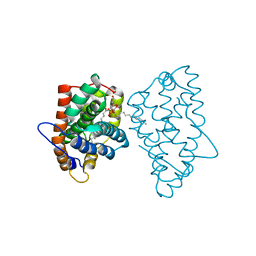 | | Crystal Structure of D48V||A47D mutant of Human Glycolipid Transfer Protein complexed with 3-O-sulfo-galactosylceramide containing nervonoyl acyl chain (24:1) | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, Glycolipid transfer protein | | Authors: | Samygina, V, Ochoa-Lizarralde, B, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2011-04-13 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
2LGI
 
 | | Atomic Resolution Protein Structures using NMR Chemical Shift Tensors | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Wylie, B.J, Sperling, L.J, Nieuwkoop, A.J, Franks, W.T, Oldfield, E, Rienstra, C.M. | | Deposit date: | 2011-07-26 | | Release date: | 2011-10-26 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Ultrahigh resolution protein structures using NMR chemical shift tensors.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4BZV
 
 | | The Solution Structure of the MLN 944-d(TACGCGTA)2 complex | | Descriptor: | 1-METHYL-9-[12-(9-METHYLPHENAZIN-10-IUM-1-YL)-12-OXO-2,11-DIAZA-5,8-DIAZONIADODEC-1-ANOYL]PHENAZIN-10-IUM, DNA | | Authors: | Serobian, A, Thomas, D.S, Ball, G.E, Denny, W.A, Wakelin, L.P.G. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of Bis(Phenazine-1-Carboxamide)-DNA Complexes: Mln 944 Binding Corrected and Extended.
Biopolymers, 101, 2014
|
|
2LEG
 
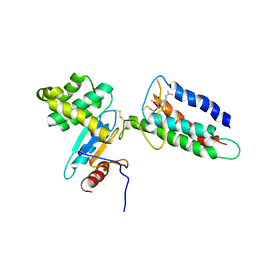 | | Membrane protein complex DsbB-DsbA structure by joint calculations with solid-state NMR and X-ray experimental data | | Descriptor: | Disulfide bond formation protein B, Thiol:disulfide interchange protein DsbA, UBIQUINONE-1, ... | | Authors: | Tang, M, Sperling, L.J, Berthold, D.A, Schwieters, C.D, Nesbitt, A.E, Nieuwkoop, A.J, Gennis, R.B, Rienstra, C.M. | | Deposit date: | 2011-06-15 | | Release date: | 2011-10-26 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution membrane protein structure by joint calculations with solid-state NMR and X-ray experimental data.
J.Biomol.Nmr, 51, 2011
|
|
2WLW
 
 | | Structure of the N-terminal capsid domain of HIV-2 | | Descriptor: | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE | | Authors: | Price, A.J, Marzetta, F, Lammers, M, Ylinen, L.M.J, Schaller, T, Wilson, S.J, Towers, G.J, James, L.C. | | Deposit date: | 2009-06-26 | | Release date: | 2009-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active Site Remodelling Switches HIV Specificity of Antiretroviral Trimcyp
Nat.Struct.Mol.Biol., 16, 2009
|
|
1FW4
 
 | |
1MZZ
 
 | | Crystal Structure of Mutant (M182T)of Nitrite Reductase | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Guo, H, Olesen, K, Xue, Y, Shapliegh, J, Sjolin, L. | | Deposit date: | 2002-10-10 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The High resolution Crystal Structures of Nitrite Reductase and its mutant Met182Thr from Rhodobacter Sphaeroides Reveal a Gating Mechanism for the Electron Transfer to the Type 1 Copper Center
To be Published
|
|
4GJQ
 
 | | Crystal structure of human GLTP bound with 12:0 monosulfatide (orthorhombic form;two subunits in asymmetric unit) | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, Glycolipid transfer protein, HEXANE | | Authors: | Cabo-Bilbao, A, Goni-de-Cerio, F, Samygina, V.R, Ochoa-Lizarralde, B, Popov, A.N, Malinina, L. | | Deposit date: | 2012-08-10 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1G2N
 
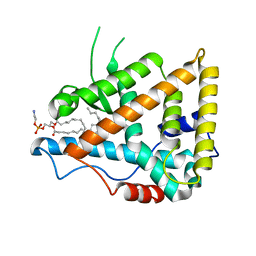 | | CRYSTAL STRUCTURE OF THE LIGAND BINDING DOMAIN OF THE ULTRASPIRACLE PROTEIN USP, THE ORTHOLOG OF RXRS IN INSECTS | | Descriptor: | L-ALPHA-PHOSPHATIDYL-BETA-OLEOYL-GAMMA-PALMITOYL-PHOSPHATIDYLETHANOLAMINE, ULTRASPIRACLE PROTEIN | | Authors: | Billas, I.M.L, Moulinier, L, Rochel, N, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-10-20 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the ligand-binding domain of the ultraspiracle protein USP, the ortholog of retinoid X receptors in insects.
J.Biol.Chem., 276, 2001
|
|
4DHS
 
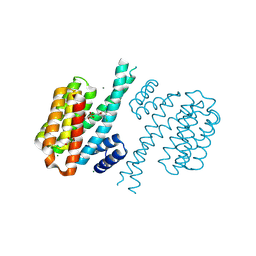 | | Small-molecule inhibitors of 14-3-3 protein-protein interactions from virtual screening | | Descriptor: | (2-{2-[(3,5-dichlorophenyl)amino]-2-oxoethoxy}phenyl)phosphonic acid, 14-3-3 PROTEIN SIGMA, CHLORIDE ION, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2012-01-30 | | Release date: | 2013-07-31 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions.
Chem.Commun.(Camb.), 49, 2013
|
|
3RWV
 
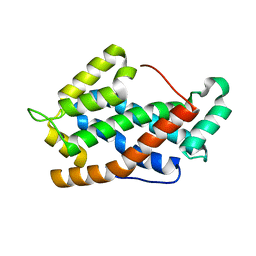 | | Crystal Structure of apo-form of Human Glycolipid Transfer Protein at 1.5 A resolution | | Descriptor: | Glycolipid transfer protein, SULFATE ION | | Authors: | Samygina, V, Cabo-Bilbao, A, Popov, A.N, Ochoa-Lizarralde, B, Goni-de-Cerio, F, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2011-05-09 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
3S0K
 
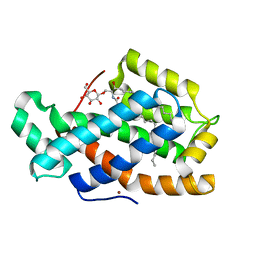 | | Crystal Structure of Human Glycolipid Transfer Protein complexed with glucosylceramide containing oleoyl acyl chain (18:1) | | Descriptor: | (9Z)-N-[(2S,3R,4E)-1-(beta-D-glucopyranosyloxy)-3-hydroxyoctadec-4-en-2-yl]octadec-9-enamide, Glycolipid transfer protein, NICKEL (II) ION | | Authors: | Cabo-Bilbao, A, Samygina, V, Popov, A.N, Ochoa-Lizarralde, B, Goni-de-Cerio, F, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2011-05-13 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
4GH0
 
 | | Crystal structure of D48V mutant of human GLTP bound with 12:0 monosulfatide | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-3-hydroxy-1-[(3-O-sulfo-beta-D-galactopyranosyl)oxy]octadec-4-en-2-yl}dodecanamide, PENTADECANE | | Authors: | Samygina, V.R, Cabo-Bilbao, A, Ochoa-Lizarralde, B, Popov, A.N, Malinina, L. | | Deposit date: | 2012-08-07 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4DHN
 
 | | Small-molecule inhibitors of 14-3-3 protein-protein interactions from virtual screening | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2012-01-30 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions.
Chem.Commun.(Camb.), 49, 2013
|
|
4DHR
 
 | | Small-molecule inhibitors of 14-3-3 protein-protein interactions from virtual screening | | Descriptor: | (2-{2-[(2-chlorophenyl)amino]-2-oxoethoxy}phenyl)phosphonic acid, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2012-01-30 | | Release date: | 2013-07-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions.
Chem.Commun.(Camb.), 49, 2013
|
|
3S0I
 
 | | Crystal Structure of D48V mutant of Human Glycolipid Transfer Protein complexed with 3-O-sulfo galactosylceramide containing nervonoyl acyl chain | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, Glycolipid transfer protein | | Authors: | Samygina, V, Popov, A.N, Cabo-Bilbao, A, Ochoa-Lizarralde, B, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2011-05-13 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
4DHQ
 
 | | Small-molecule inhibitors of 14-3-3 protein-protein interactions from virtual screening | | Descriptor: | (2-{2-[(3-chlorophenyl)amino]-2-oxoethoxy}phenyl)phosphonic acid, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2012-01-30 | | Release date: | 2013-07-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions.
Chem.Commun.(Camb.), 49, 2013
|
|
4DHO
 
 | | Small-molecule inhibitors of 14-3-3 protein-protein interactions from virtual screening | | Descriptor: | (2-{2-[(3-methoxyphenyl)amino]-2-oxoethoxy}phenyl)phosphonic acid, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2012-01-30 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions.
Chem.Commun.(Camb.), 49, 2013
|
|
