8GPN
 
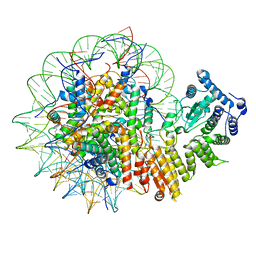 | | Human menin in complex with H3K79Me2 nucleosome | | Descriptor: | DNA (177-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Lin, J, Yu, D, Lam, W.H, Dang, S, Zhai, Y, Li, X.D. | | Deposit date: | 2022-08-26 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Menin "reads" H3K79me2 mark in a nucleosomal context.
Science, 379, 2023
|
|
4WQF
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with elongation factor G and fusidic acid in the post-translocational state | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Lin, J, Gagnon, M.G, Steitz, T.A. | | Deposit date: | 2014-10-21 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational Changes of Elongation Factor G on the Ribosome during tRNA Translocation.
Cell, 160, 2015
|
|
4WPO
 
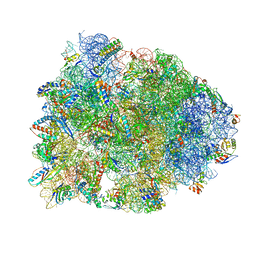 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with elongation factor G in the pre-translocational state | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Lin, J, Gagnon, M.G, Steitz, T.A. | | Deposit date: | 2014-10-20 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational Changes of Elongation Factor G on the Ribosome during tRNA Translocation.
Cell, 160, 2015
|
|
4WQU
 
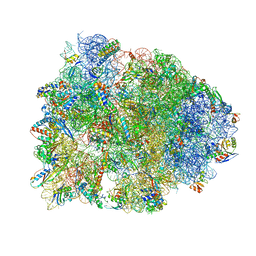 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with elongation factor G trapped by the antibiotic dityromycin | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Lin, J, Gagnon, M.G, Steitz, T.A. | | Deposit date: | 2014-10-22 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational Changes of Elongation Factor G on the Ribosome during tRNA Translocation.
Cell, 160, 2015
|
|
4WQY
 
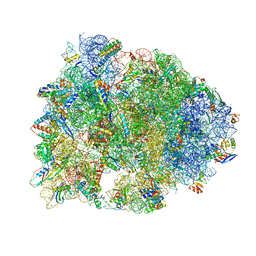 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with elongation factor G in the post-translocational state (without fusitic acid) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Lin, J, Gagnon, M.G, Steitz, T.A. | | Deposit date: | 2014-10-22 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational Changes of Elongation Factor G on the Ribosome during tRNA Translocation.
Cell, 160, 2015
|
|
3J3P
 
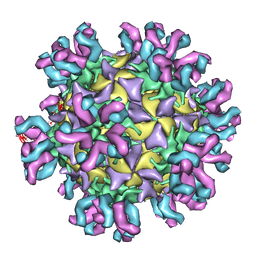 | | Conformational Shift of a Major Poliovirus Antigen Confirmed by Immuno-Cryogenic Electron Microscopy: 135S Poliovirus and C3-Fab Complex | | Descriptor: | C3 antibody, heavy chain, light chain, ... | | Authors: | Lin, J, Cheng, N, Hogle, J.M, Steven, A.C, Belnap, D.M. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Conformational shift of a major poliovirus antigen confirmed by immuno-cryogenic electron microscopy.
J.Immunol., 191, 2013
|
|
3J3O
 
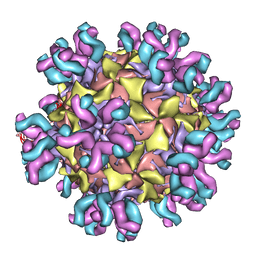 | | Conformational Shift of a Major Poliovirus Antigen Confirmed by Immuno-Cryogenic Electron Microscopy: 160S Poliovirus and C3-Fab Complex | | Descriptor: | C3 antibody, heavy chain, light chain, ... | | Authors: | Lin, J, Cheng, N, Hogle, J.M, Steven, A.C, Belnap, D.M. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.1 Å) | | Cite: | Conformational shift of a major poliovirus antigen confirmed by immuno-cryogenic electron microscopy.
J.Immunol., 191, 2013
|
|
3J67
 
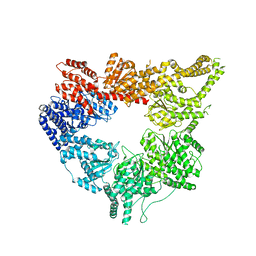 | | Structural mechanism of the dynein powerstroke (post-powerstroke state) | | Descriptor: | Dynein motor domain | | Authors: | Lin, J, Okada, K, Raytchev, M, Smith, M.C, Nicastro, D. | | Deposit date: | 2013-12-22 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (34 Å) | | Cite: | Structural mechanism of the dynein power stroke.
Nat.Cell Biol., 16, 2014
|
|
3J68
 
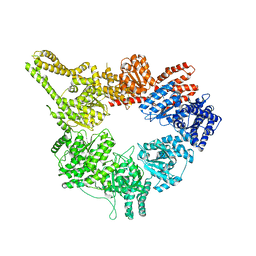 | | Structural mechanism of the dynein powerstroke (pre-powerstroke state) | | Descriptor: | Dynein motor domain | | Authors: | Lin, J, Okada, K, Raytchev, M, Smith, M.C, Nicastro, D. | | Deposit date: | 2013-12-23 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (30 Å) | | Cite: | Structural mechanism of the dynein power stroke.
Nat.Cell Biol., 16, 2014
|
|
1ZOM
 
 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in complex with a peptidomimetic Inhibitor | | Descriptor: | (S)-2-(3-((R)-1-(4-BROMOPHENYL)ETHYL)UREIDO)-N-((S)-1-((S)-5-GUANIDINO-1-OXO-1-(THIAZOL-2-YL)PENTAN-2-YLAMINO)-3-METHYL-1-OXOBUTAN-2-YL)-5-UREIDOPENTANAMIDE, Coagulation factor XI, SULFATE ION | | Authors: | Lin, J, Deng, H, Jin, L, Pandey, P, Rynkiewicz, M, Bibbins, F, Cantin, S, Quinn, J, Magee, S, Gorga, J. | | Deposit date: | 2005-05-13 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Design, synthesis, and biological evaluation of peptidomimetic inhibitors of factor XIa as novel anticoagulants.
J.Med.Chem., 49, 2006
|
|
1ZPZ
 
 | | Factor XI catalytic domain complexed with N-((R)-1-(4-bromophenyl)ethyl)urea-Asn-Val-Arg-alpha-ketothiazole | | Descriptor: | Coagulation factor XI, N~2~-({[(1R)-1-(4-BROMOPHENYL)ETHYL]AMINO}CARBONYL)ASPARAGINYL-N~1~-{4-{[AMINO(IMINO)METHYL]AMINO}-1-[2,3-DIHYDRO-1,3-THIAZOL-2-YL(HYDROXY)METHYL]BUTYL}VALINAMIDE, SULFATE ION | | Authors: | Lin, J, Deng, H, Jin, L, Prandey, P, Rynkiewicz, M.J, Bibbins, F, Cantin, S, Quinn, J, Magee, S, Gorga, J. | | Deposit date: | 2005-05-18 | | Release date: | 2006-05-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, Synthesis and Biological Evaluation of Peptidomimetic FXIa Inhibitors
To be Published
|
|
1TUM
 
 | | MUTT PYROPHOSPHOHYDROLASE-METAL-NUCLEOTIDE-METAL COMPLEX, NMR, 16 STRUCTURES | | Descriptor: | COBALT TETRAAMMINE ION, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, ... | | Authors: | Lin, J, Abeygunawardana, C, Frick, D.N, Bessman, M.J, Mildvan, A.S. | | Deposit date: | 1996-12-05 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the quaternary MutT-M2+-AMPCPP-M2+ complex and mechanism of its pyrophosphohydrolase action.
Biochemistry, 36, 1997
|
|
5E1K
 
 | | Selenomethionine Ca2+-Calmodulin from Paramecium tetraurelia SAD data | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Calmodulin | | Authors: | Lin, J, van den Bedem, H, Brunger, A.T, Wilson, M.A. | | Deposit date: | 2015-09-29 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic resolution experimental phase information reveals extensive disorder and bound 2-methyl-2,4-pentanediol in Ca(2+)-calmodulin.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5E1P
 
 | | Ca(2+)-Calmodulin from Paramecium tetraurelia qFit disorder model | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Calmodulin | | Authors: | Lin, J, van den Bedem, H, Brunger, A.T, Wilson, M.A. | | Deposit date: | 2015-09-29 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Atomic resolution experimental phase information reveals extensive disorder and bound 2-methyl-2,4-pentanediol in Ca(2+)-calmodulin.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5E1N
 
 | | Selenomethionine Ca2+-Calmodulin from Paramecium tetraurelia qFit disorder model | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Calmodulin | | Authors: | Lin, J, van den Bedem, H, Brunger, A.T, Wilson, M.A. | | Deposit date: | 2015-09-29 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic resolution experimental phase information reveals extensive disorder and bound 2-methyl-2,4-pentanediol in Ca(2+)-calmodulin.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4M5D
 
 | |
3TBN
 
 | |
3TBM
 
 | | Crystal structure of a type 4 CDGSH iron-sulfur protein. | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, L(+)-TARTARIC ACID, NONAETHYLENE GLYCOL, ... | | Authors: | Lin, J, Zhang, L, Ye, K. | | Deposit date: | 2011-08-07 | | Release date: | 2011-10-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Structure and Molecular Evolution of CDGSH Iron-Sulfur Domains.
Plos One, 6, 2011
|
|
3TBO
 
 | |
4LEB
 
 | |
4LE8
 
 | |
4LEE
 
 | | Structure of the Als3 adhesin from Candida albicans, residues 1-313 (mature sequence), triple mutant in the binding cavity: K59M, A116V, Y301F | | Descriptor: | Agglutinin-like protein 3 | | Authors: | Lin, J, Garnett, J.A, Cota, E. | | Deposit date: | 2013-06-25 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Peptide-binding Cavity Is Essential for Als3-mediated Adhesion of Candida albicans to Human Cells.
J.Biol.Chem., 289, 2014
|
|
3PLA
 
 | | Crystal structure of a catalytically active substrate-bound box C/D RNP from Sulfolobus solfataricus | | Descriptor: | 50S ribosomal protein L7Ae, C/D guide RNA, Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase, ... | | Authors: | Lin, J, Lai, S, Jia, R, Xu, A, Zhang, L, Lu, J, Ye, K. | | Deposit date: | 2010-11-15 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis for site-specific ribose methylation by box C/D RNA protein complexes.
Nature, 469, 2011
|
|
3QOU
 
 | | Crystal Structure of E. coli YbbN | | Descriptor: | CALCIUM ION, protein ybbN | | Authors: | Lin, J, Wilson, M.A. | | Deposit date: | 2011-02-10 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Escherichia coli Thioredoxin-like Protein YbbN Contains an Atypical Tetratricopeptide Repeat Motif and Is a Negative Regulator of GroEL.
J.Biol.Chem., 286, 2011
|
|
2L2O
 
 | |
