1YT1
 
 | | Crystal Structure of the Unliganded Form of GRP94, the ER Hsp90: Basis for Nucleotide-Induced Conformational Change, GRP94N(DELTA)41 APO CRYSTAL | | Descriptor: | Endoplasmin, PENTAETHYLENE GLYCOL, TETRAETHYLENE GLYCOL | | Authors: | Dollins, D.E, Immormino, R.M, Gewirth, D.T. | | Deposit date: | 2005-02-09 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Unliganded GRP94, the Endoplasmic Reticulum Hsp90: BASIS FOR NUCLEOTIDE-INDUCED CONFORMATIONAL CHANGE
J.Biol.Chem., 280, 2005
|
|
7AIK
 
 | | Ribonucleotide Reductase R2 protein from Aquifex aeolicus | | Descriptor: | FE (II) ION, Ribonucleoside-diphosphate reductase subunit beta,Ribonucleoside-diphosphate reductase subunit beta | | Authors: | Rehling, D, Scaletti, E.R, Stenmark, P. | | Deposit date: | 2020-09-27 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Biochemical Investigation of Class I Ribonucleotide Reductase from the Hyperthermophile Aquifex aeolicus.
Biochemistry, 61, 2022
|
|
8S6J
 
 | | NavMs in complex with riluzole | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 6-(trifluoromethoxy)-1,3-benzothiazol-2-amine, CHLORIDE ION, ... | | Authors: | Hollingworth, D, Sula, A, Mykhaylyk, V, Wallace, B.A. | | Deposit date: | 2024-02-27 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for the rescue of hyperexcitable cells by the amyotrophic lateral sclerosis drug Riluzole.
Nat Commun, 15, 2024
|
|
1YSZ
 
 | | Crystal Structure of the Unliganded Form of GRP94, the ER Hsp90: Basis for Nucleotide-Induced Conformational Change, GRP94N(DELTA)41 APO CRYSTAL SOAKED WITH NECA | | Descriptor: | Endoplasmin, N-ETHYL-5'-CARBOXAMIDO ADENOSINE, TETRAETHYLENE GLYCOL | | Authors: | Dollins, D.E, Immormino, R.M, Gewirth, D.T. | | Deposit date: | 2005-02-09 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of Unliganded GRP94, the Endoplasmic Reticulum Hsp90: BASIS FOR NUCLEOTIDE-INDUCED CONFORMATIONAL CHANGE
J.Biol.Chem., 280, 2005
|
|
5T7Z
 
 | | Monoclinic crystal form of the EpoB NRPS cyclization-docking bidomain from Sorangium cellulosum | | Descriptor: | EpoB | | Authors: | Dowling, D.P, Kung, Y, Croft, A.K, Taghizadeh, K, Kelly, W.L, Walsh, C.T, Drennan, C.L. | | Deposit date: | 2016-09-06 | | Release date: | 2016-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural elements of an NRPS cyclization domain and its intermodule docking domain.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3EW8
 
 | | Crystal Structure Analysis of human HDAC8 D101L variant | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Dowling, D.P, Gantt, S.L, Gattis, S.G, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2008-10-14 | | Release date: | 2008-12-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of human histone deacetylase 8 and its site-specific variants complexed with substrate and inhibitors.
Biochemistry, 47, 2008
|
|
2NUM
 
 | | Soluble domain of Rieske Iron-Sulfur Protein | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ubiquinol-cytochrome c reductase iron-sulfur subunit | | Authors: | Kolling, D, Brunzelle, J, Lhee, S, Crofts, A.R, Nair, S.K. | | Deposit date: | 2006-11-09 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Atomic resolution structures of rieske iron-sulfur protein: role of hydrogen bonds in tuning the redox potential of iron-sulfur clusters.
Structure, 15, 2007
|
|
2NVG
 
 | | Soluble domain of Rieske Iron Sulfur protein. | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, Ubiquinol-cytochrome c reductase iron-sulfur subunit | | Authors: | Kolling, D, Brunzelle, J, Lhee, S, Crofts, A.R, Nair, S.K. | | Deposit date: | 2006-11-12 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Atomic resolution structures of rieske iron-sulfur protein: role of hydrogen bonds in tuning the redox potential of iron-sulfur clusters.
Structure, 15, 2007
|
|
6FNL
 
 | | Crystal Structure of Ephrin B4 (EphB4) Receptor Protein Kinase | | Descriptor: | Ephrin type-B receptor 4 | | Authors: | Kudlinzki, D, Troester, A, Witt, K, Linhard, V.L, Saxena, K, Schwalbe, H. | | Deposit date: | 2018-02-04 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.269 Å) | | Cite: | NVP-BHG712: Effects of Regioisomers on the Affinity and Selectivity toward the EPHrin Family.
ChemMedChem, 13, 2018
|
|
6FNI
 
 | | Crystal Structure of Ephrin B4 (EphB4) Receptor Protein Kinase with NVP-BHG712 | | Descriptor: | 4-methyl-3-[(1-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-yl)amino]-~{N}-[3-(trifluoromethyl)phenyl]benzamide, Ephrin type-B receptor 4 | | Authors: | Kudlinzki, D, Troester, A, Witt, K, Linhard, V.L, Saxena, K, Schwalbe, H. | | Deposit date: | 2018-02-04 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.468 Å) | | Cite: | NVP-BHG712: Effects of Regioisomers on the Affinity and Selectivity toward the EPHrin Family.
ChemMedChem, 13, 2018
|
|
6FNM
 
 | | Crystal Structure of Ephrin B4 (EphB4) Receptor Protein Kinase with Dasatinib | | Descriptor: | Ephrin type-B receptor 4, N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE | | Authors: | Kudlinzki, D, Troester, A, Witt, K, Linhard, V.L, Saxena, K, Schwalbe, H. | | Deposit date: | 2018-02-04 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.157 Å) | | Cite: | NVP-BHG712: Effects of Regioisomers on the Affinity and Selectivity toward the EPHrin Family.
ChemMedChem, 13, 2018
|
|
5T81
 
 | | Rhombohedral crystal form of the EpoB NRPS cyclization-docking bidomain from Sorangium cellulosum | | Descriptor: | EpoB, GLYCEROL | | Authors: | Dowling, D.P, Kung, Y, Croft, A.K, Taghizadeh, K, Kelly, W.L, Walsh, C.T, Drennan, C.L. | | Deposit date: | 2016-09-06 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Structural elements of an NRPS cyclization domain and its intermodule docking domain.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6FNF
 
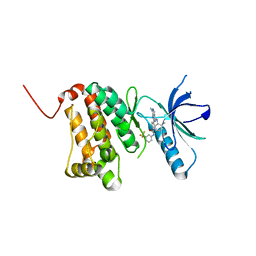 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with NVP-BHG712 | | Descriptor: | 4-methyl-3-[(1-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-yl)amino]-~{N}-[3-(trifluoromethyl)phenyl]benzamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Troester, A, Witt, K, Linhard, V.L, Gande, S.L, Saxena, K, Schwalbe, H. | | Deposit date: | 2018-02-04 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.556 Å) | | Cite: | NVP-BHG712: Effects of Regioisomers on the Affinity and Selectivity toward the EPHrin Family.
ChemMedChem, 13, 2018
|
|
1OZS
 
 | |
4YMG
 
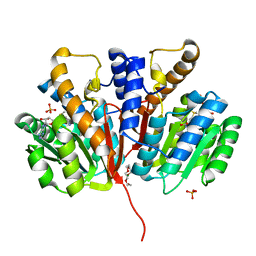 | | Crystal structure of SAM-bound Podospora anserina methyltransferase PaMTH1 | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Putative SAM-dependent O-methyltranferase, ... | | Authors: | Kudlinzki, D, Linhard, V.L, Chatterjee, D, Saxena, K, Sreeramulu, S, Schwalbe, H. | | Deposit date: | 2015-03-06 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structure and Biophysical Characterization of the S-Adenosylmethionine-dependent O-Methyltransferase PaMTH1, a Putative Enzyme Accumulating during Senescence of Podospora anserina.
J.Biol.Chem., 290, 2015
|
|
1YT0
 
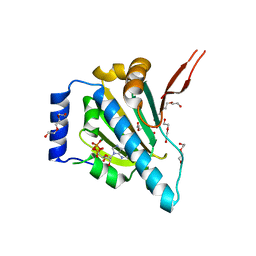 | | Crystal Structure of the Unliganded Form of GRP94, the ER Hsp90: Basis for Nucleotide-Induced Conformational Change, GRP94N(DELTA)41 APO CRYSTAL SOAKED WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Endoplasmin, MAGNESIUM ION, ... | | Authors: | Dollins, D.E, Immormino, R.M, Gewirth, D.T. | | Deposit date: | 2005-02-09 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Unliganded GRP94, the Endoplasmic Reticulum Hsp90: BASIS FOR NUCLEOTIDE-INDUCED CONFORMATIONAL CHANGE
J.Biol.Chem., 280, 2005
|
|
1YT2
 
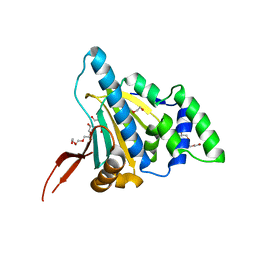 | | Crystal Structure of the Unliganded Form of GRP94, the ER Hsp90: Basis for Nucleotide-Induced Conformational Change, GRP94N APO CRYSTAL | | Descriptor: | Endoplasmin, TETRAETHYLENE GLYCOL | | Authors: | Dollins, D.E, Immormino, R.M, Gewirth, D.T. | | Deposit date: | 2005-02-09 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of Unliganded GRP94, the Endoplasmic Reticulum Hsp90: BASIS FOR NUCLEOTIDE-INDUCED CONFORMATIONAL CHANGE
J.Biol.Chem., 280, 2005
|
|
2NUK
 
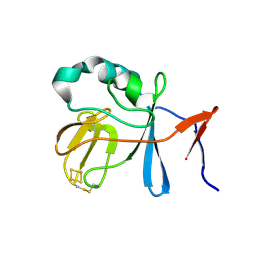 | | Soluble Domain of the Rieske Iron-Sulfur Protein from Rhodobacter sphaeroides | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ubiquinol-cytochrome c reductase iron-sulfur subunit | | Authors: | Kolling, D, Brunzelle, J, Lhee, S, Crofts, A.R, Nair, S.K. | | Deposit date: | 2006-11-09 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomic resolution structures of rieske iron-sulfur protein: role of hydrogen bonds in tuning the redox potential of iron-sulfur clusters.
Structure, 15, 2007
|
|
3I4U
 
 | |
7KIR
 
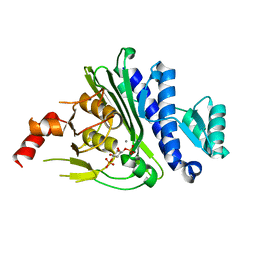 | | Crystal structure of inositol polyphosphate 1-phosphatase (INPP1) D54A mutant in complex with inositol (1,4)-bisphosphate | | Descriptor: | CALCIUM ION, D-MYO-INOSITOL-1,4-BISPHOSPHATE, Inositol polyphosphate 1-phosphatase | | Authors: | Dollins, D.E, Xiong, J.-P, Ren, Y, York, J.D. | | Deposit date: | 2020-10-24 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A structural basis for lithium and substrate binding of an inositide phosphatase.
J.Biol.Chem., 296, 2020
|
|
3MZ3
 
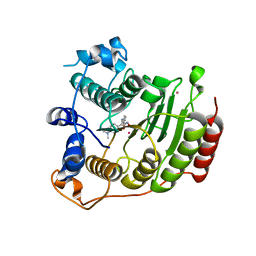 | | Crystal structure of Co2+ HDAC8 complexed with M344 | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, COBALT (II) ION, Histone deacetylase 8, ... | | Authors: | Dowling, D.P, Gattis, S.G, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2010-05-11 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of metal-substituted human histone deacetylase 8 provide mechanistic inferences on biological function.
Biochemistry, 49, 2010
|
|
4MDZ
 
 | | Crystal structure of a HD-GYP domain (a cyclic-di-GMP phosphodiesterase) containing a tri-nuclear metal centre | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), FE (III) ION, Metal dependent phosphohydrolase, ... | | Authors: | Bellini, D, Walsh, M.A, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2013-08-23 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure of an HD-GYP domain cyclic-di-GMP phosphodiesterase reveals an enzyme with a novel trinuclear catalytic iron centre.
Mol.Microbiol., 91, 2014
|
|
3MZ4
 
 | | Crystal structure of D101L Mn2+ HDAC8 complexed with M344 | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, GLYCEROL, Histone deacetylase 8, ... | | Authors: | Dowling, D.P, Gattis, S.G, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2010-05-11 | | Release date: | 2010-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Structures of metal-substituted human histone deacetylase 8 provide mechanistic inferences on biological function.
Biochemistry, 49, 2010
|
|
7AT9
 
 | | Structure of protein kinase ck2 catalytic subunit (csnk2a2 gene product) in complex with the ATP-competitive inhibitor MB002 and the alphaD-pocket ligand 3,4-dichlorophenethylamine | | Descriptor: | 1,2-ETHANEDIOL, 2-(3,4-dichlorophenyl)ethanamine, 3-(4,5,6,7-tetrabromo-1H-benzotriazol-1-yl)propan-1-ol, ... | | Authors: | Lindenblatt, D, Applegate, V, Nickelsen, A, Klussmann, M, Neundorf, I, Goetz, C, Jose, J, Niefind, K. | | Deposit date: | 2020-10-29 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Molecular Plasticity of Crystalline CK2 alpha ' Leads to KN2, a Bivalent Inhibitor of Protein Kinase CK2 with Extraordinary Selectivity.
J.Med.Chem., 65, 2022
|
|
7AT5
 
 | | Structure of protein kinase ck2 catalytic subunit (csnk2a1 gene product) in complex with the bivalent inhibitor KN2 | | Descriptor: | 1,2-ETHANEDIOL, 2-(3,4-dichlorophenyl)ethanamine, Casein kinase II subunit alpha, ... | | Authors: | Lindenblatt, D, Applegate, V, Nickelsen, A, Klussmann, M, Neundorf, I, Goetz, C, Jose, J, Niefind, K. | | Deposit date: | 2020-10-29 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Molecular Plasticity of Crystalline CK2 alpha ' Leads to KN2, a Bivalent Inhibitor of Protein Kinase CK2 with Extraordinary Selectivity.
J.Med.Chem., 65, 2022
|
|
