4QPC
 
 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (Y200A) from zebrafish | | Descriptor: | 10-formyltetrahydrofolate dehydrogenase | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-06-23 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4TT8
 
 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (wild-type) complex with 10-formyl-5,8-dideazafolate | | Descriptor: | 10-formyltetrahydrofolate dehydrogenase, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-(4-{[(2-amino-4-hydroxyquinazolin-6-yl)methyl](formyl)amino}benzoyl)-L-glutamic acid | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-06-20 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4TTS
 
 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (Y200A) complex with 10-formyl-5,8-dideazafolate | | Descriptor: | 10-formyltetrahydrofolate dehydrogenase, N-(4-{[(2-amino-4-hydroxyquinazolin-6-yl)methyl](formyl)amino}benzoyl)-L-glutamic acid | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-06-23 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4TS4
 
 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (wild-type) from zebrafish | | Descriptor: | 10-formyltetrahydrofolate dehydrogenase | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-06-18 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4QPD
 
 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (wild-type) complex with tetrahydrofolate | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, 10-formyltetrahydrofolate dehydrogenase, DI(HYDROXYETHYL)ETHER | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-06-23 | | Release date: | 2015-04-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R8V
 
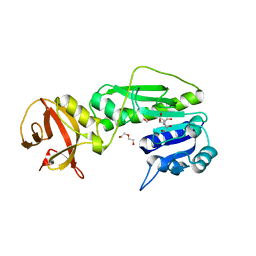 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (wild-type) complex with formate | | Descriptor: | 10-formyltetrahydrofolate dehydrogenase, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-09-03 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7EI3
 
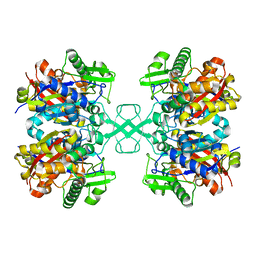 | |
7EI4
 
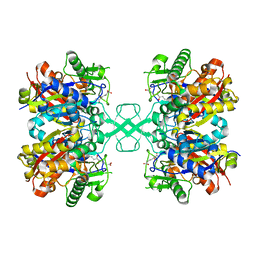 | | Crystal structure of MasL in complex with a novel covalent inhibitor, collimonin C | | Descriptor: | (6S,7R,9E)-6,7-bis(oxidanyl)hexadeca-9,15-dien-11,13-diynoic acid, Acetyl-CoA C-acyltransferase | | Authors: | Lin, C.C, Huang, K.F, Yang, Y.L. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Integrated omics approach to unveil antifungal bacterial polyynes as acetyl-CoA acetyltransferase inhibitors.
Commun Biol, 5, 2022
|
|
5GZ4
 
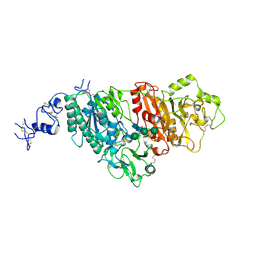 | |
5GZ5
 
 | |
7FEA
 
 | | PY14 in complex with Col-D | | Descriptor: | (6~{R},7~{R},9~{E})-6,7-bis(oxidanyl)hexadeca-9,15-dien-11,13-diynoic acid, Acetyl-CoA C-acyltransferase | | Authors: | Lin, C.C, Ko, T.P, Huang, K.F, Yang, Y.L. | | Deposit date: | 2021-07-19 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Integrated omics approach to unveil antifungal bacterial polyynes as acetyl-CoA acetyltransferase inhibitors.
Commun Biol, 5, 2022
|
|
4OM5
 
 | | Crystal structure of CTX A4 from Taiwan Cobra (Naja naja atra) | | Descriptor: | Cytotoxin 4 | | Authors: | Lin, C.C, Chang, C.I, Wu, W.G. | | Deposit date: | 2014-01-26 | | Release date: | 2014-06-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Endocytotic Routes of Cobra Cardiotoxins Depend on Spatial Distribution of Positively Charged and Hydrophobic Domains to Target Distinct Types of Sulfated Glycoconjugates on Cell Surface.
J.Biol.Chem., 289, 2014
|
|
4OM4
 
 | | Crystal structure of CTX A2 from Taiwan Cobra (Naja naja atra) | | Descriptor: | Cytotoxin 2 | | Authors: | Lin, C.C, Chang, C.I, Wu, W.G. | | Deposit date: | 2014-01-26 | | Release date: | 2014-06-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Endocytotic Routes of Cobra Cardiotoxins Depend on Spatial Distribution of Positively Charged and Hydrophobic Domains to Target Distinct Types of Sulfated Glycoconjugates on Cell Surface.
J.Biol.Chem., 289, 2014
|
|
7C1I
 
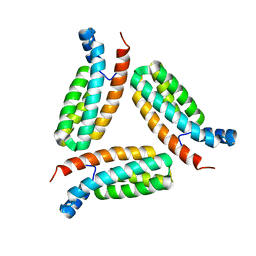 | | Crystal structure of histidine-containing phosphotransfer protein B (HptB) from Pseudomonas aeruginosa PAO1 | | Descriptor: | Histidine kinase | | Authors: | Chen, S.K, Guan, H.H, Wu, P.H, Lin, L.T, Wu, M.C, Chang, H.Y, Chen, N.C, Lin, C.C, Chuankhayan, P, Huang, Y.C, Lin, P.J, Chen, C.J. | | Deposit date: | 2020-05-04 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural insights into the histidine-containing phospho-transfer protein and receiver domain of sensor histidine kinase suggest a complex model in the two-component regulatory system in Pseudomonas aeruginosa
Iucrj, 7, 2020
|
|
7C1J
 
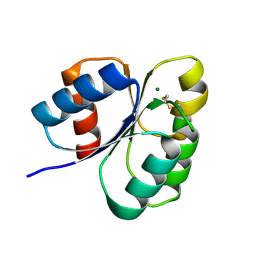 | | Crystal structure of the receiver domain of sensor histidine kinase PA1611 (PA1611REC) from Pseudomonas aeruginosa PAO1 with magnesium ion coordinated in the active site cleft | | Descriptor: | Histidine kinase, MAGNESIUM ION | | Authors: | Chen, S.K, Guan, H.H, Wu, P.H, Lin, L.T, Wu, M.C, Chang, H.Y, Chen, N.C, Lin, C.C, Chuankhayan, P, Huang, Y.C, Lin, P.J, Chen, C.J. | | Deposit date: | 2020-05-04 | | Release date: | 2020-11-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insights into the histidine-containing phospho-transfer protein and receiver domain of sensor histidine kinase suggest a complex model in the two-component regulatory system in Pseudomonas aeruginosa
Iucrj, 7, 2020
|
|
5Z2G
 
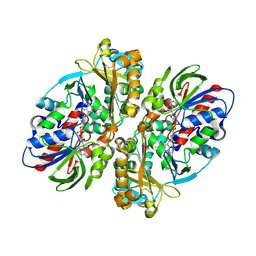 | | Crystal Structure of L-amino acid oxidase from venom of Naja atra | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, L-amino acid oxidase | | Authors: | Kumar, J.V, Chien, K.Y, Wu, W.G, Lin, C.C, Chiang, L.C, Lin, T.H. | | Deposit date: | 2018-01-02 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.676 Å) | | Cite: | Crystal Structure of L-amino acid oxidase from naja atra (Taiwan Cobra)
To Be Published
|
|
6AB5
 
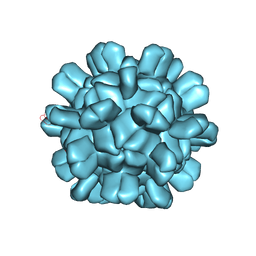 | | Cryo-EM structure of T=1 Penaeus vannamei nodavirus | | Descriptor: | Capsid protein | | Authors: | Chen, N.C, Miyazaki, N, Yoshimura, M, Guan, H.H, Lin, C.C, Iwasaki, K, Chen, C.J. | | Deposit date: | 2018-07-20 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
6AB6
 
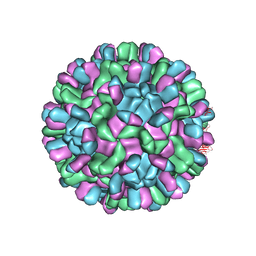 | | Cryo-EM structure of T=3 Penaeus vannamei nodavirus | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Chen, N.C, Miyazaki, N, Yoshimura, M, Guan, H.H, Lin, C.C, Iwasaki, K, Chen, C.J. | | Deposit date: | 2018-07-20 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
5YL0
 
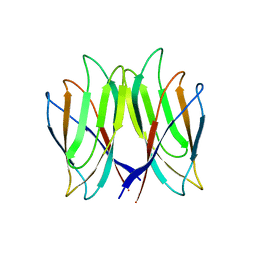 | | The crystal structure of Penaeus vannamei nodavirus P-domain (P212121) | | Descriptor: | Capsid protein | | Authors: | Chen, N.C, Yoshimura, M, Lin, C.C, Guan, H.H, Chuankhayan, P, Chen, C.J. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
5YKX
 
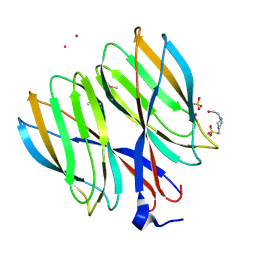 | | The crystal structure of Macrobrachium rosenbergii nodavirus P-domain with Cd ion | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CADMIUM ION, Capsid protein, ... | | Authors: | Chen, N.C, Yoshimura, M, Lin, C.C, Guan, H.H, Chuankhayan, P, Chen, C.J. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2019-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
5YKZ
 
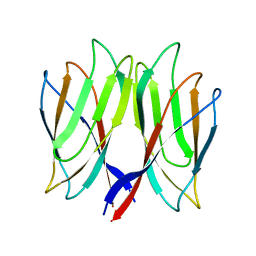 | | The crystal structure of Penaeus vannamei nodavirus P-domain (P21) | | Descriptor: | Capsid protein | | Authors: | Chen, N.C, Yoshimura, M, Lin, C.C, Guan, H.H, Chuankhayan, P, Chen, C.J. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
5YKV
 
 | | The crystal structure of Macrobrachium rosenbergii nodavirus P-domain | | Descriptor: | Capsid protein | | Authors: | Chen, N.C, Yoshimura, M, Lin, C.C, Guan, H.H, Chuankhayan, P, Chen, C.J. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
5YKU
 
 | | The crystal structure of Macrobrachium rosenbergii nodavirus P-domain with Zn ions | | Descriptor: | Capsid protein, ZINC ION | | Authors: | Chen, N.C, Yoshimura, M, Lin, C.C, Guan, H.H, Chuankhayan, P, Chen, C.J. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
5YL1
 
 | | T=1 subviral particle of Penaeus vannamei nodavirus capsid protein deletion mutant (delta 1-37 & 251-368) | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Chen, N.C, Yoshimura, M, Lin, C.C, Guan, H.H, Chuankhayan, P, Chen, C.J. | | Deposit date: | 2017-10-16 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
8SHH
 
 | | Crystal structure of EvdS6 decarboxylase in ligand free state | | Descriptor: | DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, dTDP-glucose 4,6-dehydratase | | Authors: | Sharma, P, Frigo, L, Dulin, C.C, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2023-04-14 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | EvdS6 is a bifunctional decarboxylase from the everninomicin gene cluster.
J.Biol.Chem., 299, 2023
|
|
