2YOA
 
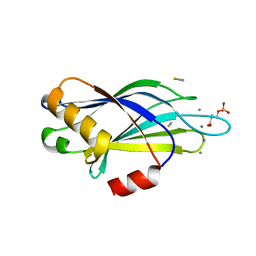 | | Synaptotagmin-1 C2B domain with phosphoserine | | Descriptor: | CALCIUM ION, PHOSPHOSERINE, SYNAPTOTAGMIN-1, ... | | Authors: | Honigmann, A, van den Bogaart, G, Iraheta, E, Risselada, H.J, Milovanovic, D, Mueller, V, Muellar, S, Diederichsen, U, Fasshauer, D, Grubmuller, H, Hell, S.W, Eggeling, C, Kuhnel, K, Jahn, R. | | Deposit date: | 2012-10-22 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Phosphatidylinositol 4,5-Bisphosphate Clusters Act as Molecular Beacons for Vesicle Recruitment
Nat.Struct.Mol.Biol., 20, 2013
|
|
4GFT
 
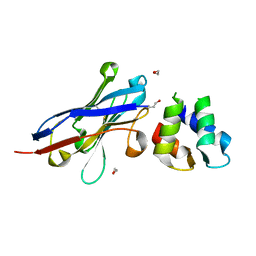 | | Malaria invasion machinery protein-Nanobody complex | | Descriptor: | 1,2-ETHANEDIOL, Myosin A tail domain interacting protein, Nanobody | | Authors: | Khamrui, S, Turley, S, Pardon, E, Steyaert, J, Verlinde, C, Fan, E, Bergman, L.W, Hol, W.G.J. | | Deposit date: | 2012-08-03 | | Release date: | 2013-07-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of the D3 domain of Plasmodium falciparum myosin tail interacting protein MTIP in complex with a nanobody.
Mol.Biochem.Parasitol., 190, 2013
|
|
4AU8
 
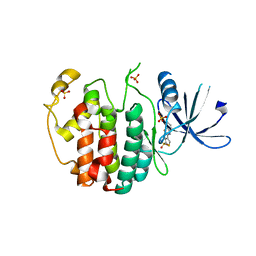 | | Crystal structure of compound 4a in complex with cdk5, showing an unusual binding mode to the hinge region via a water molecule | | Descriptor: | 4-(1,3-benzothiazol-2-yl)thiophene-2-sulfonamide, CYCLIN-DEPENDENT KINASE 5, IMIDAZOLE, ... | | Authors: | Malmstrom, J, Viklund, J, Slivo, C, Costa, A, Maudet, M, Sandelin, C, Hiller, G, Olsson, L.L, Aagaard, A, Geschwindner, S, Xue, Y, Vasange, M. | | Deposit date: | 2012-05-14 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and Structure-Activity Relationship of 4-(1,3-Benzothiazol-2-Yl)-Thiophene-2-Sulfonamides as Cyclin-Dependent Kinase 5 (Cdk5)/P25 Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1G27
 
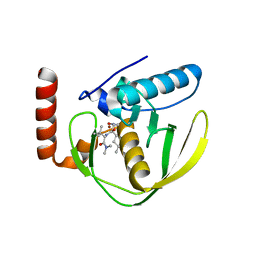 | | CRYSTAL STRUCTURE OF E.COLI POLYPEPTIDE DEFORMYLASE COMPLEXED WITH THE INHIBITOR BB-3497 | | Descriptor: | 2-[(FORMYL-HYDROXY-AMINO)-METHYL]-HEXANOIC ACID (1-DIMETHYLCARBAMOYL-2,2-DIMETHYL-PROPYL)-AMIDE, NICKEL (II) ION, POLYPEPTIDE DEFORMYLASE | | Authors: | Clements, J.M, Beckett, P, Brown, A, Catlin, C, Lobell, M, Palan, S, Thomas, W, Whittaker, M, Baker, P.J, Rodgers, H.F, Barynin, V, Rice, D.W, Hunter, M.G. | | Deposit date: | 2000-10-17 | | Release date: | 2001-10-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antibiotic activity and characterization of BB-3497, a novel peptide deformylase inhibitor.
Antimicrob.Agents Chemother., 45, 2001
|
|
1G2A
 
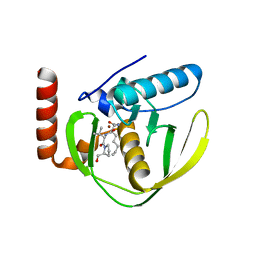 | | THE CRYSTAL STRUCTURE OF E.COLI PEPTIDE DEFORMYLASE COMPLEXED WITH ACTINONIN | | Descriptor: | ACTINONIN, NICKEL (II) ION, POLYPEPTIDE DEFORMYLASE | | Authors: | Clements, J.M, Beckett, P, Brown, A, Catlin, C, Lobell, M, Palan, S, Thomas, W, Whittaker, M, Baker, P.J, Rodgers, H.F, Barynin, V, Rice, D.W, Hunter, M.G. | | Deposit date: | 2000-10-18 | | Release date: | 2001-10-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Antibiotic activity and characterization of BB-3497, a novel peptide deformylase inhibitor.
Antimicrob.Agents Chemother., 45, 2001
|
|
2KG2
 
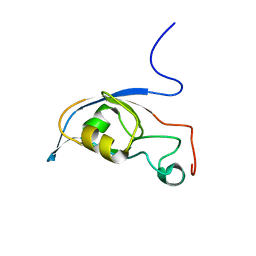 | | Solution structure of a PDZ protein | | Descriptor: | Tax1-binding protein 3 | | Authors: | Durney, M.A, Birrane, G, Anklin, C, Soni, A, Ladias, J.A.A. | | Deposit date: | 2009-03-02 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the human Tax-interacting protein-1.
J.Biomol.Nmr, 45, 2009
|
|
2MM8
 
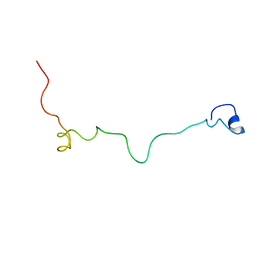 | | Structural and biochemical characterization of Jaburetox | | Descriptor: | Urease | | Authors: | Dobrovolska, O, Lopez, F, Ciurli, S, Zambelli, B, Carlini, C. | | Deposit date: | 2014-03-12 | | Release date: | 2015-01-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C, 15N resonance assignments of reduced Jaburetox
To be Published
|
|
2O20
 
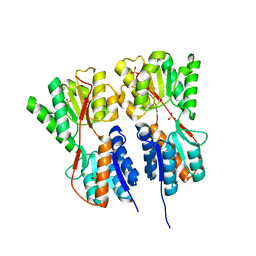 | | Crystal structure of transcription regulator CcpA of Lactococcus lactis | | Descriptor: | CHLORIDE ION, Catabolite control protein A, SULFATE ION | | Authors: | Loll, B, Kowalczyk, M, Alings, C, Chieduch, A, Bardowski, J, Saenger, W, Biesiadka, J. | | Deposit date: | 2006-11-29 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the transcription regulator CcpA from Lactococcus lactis
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2NXU
 
 | | Atomic structure of translation initiation factor aIF2 beta-subunit from Archaebacteria sulfolobus solfataricus: high resolution NMR in solution | | Descriptor: | Translation initiation factor 2 beta subunit | | Authors: | Vasile, F, Pechkova, E, Stolboushkina, E, Garber, M, Nicolini, C. | | Deposit date: | 2006-11-20 | | Release date: | 2007-10-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the beta-subunit of the translation initiation factor aIF2 from archaebacteria Sulfolobus solfataricus.
Proteins, 70, 2007
|
|
