6YEF
 
 | | 70S initiation complex with assigned rRNA modifications from Staphylococcus aureus | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S11, ... | | Authors: | Fatkhullin, B, Golubev, A, Khusainov, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2020-03-24 | | Release date: | 2021-01-27 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the ribosome functional complex of the human pathogen Staphylococcus aureus at 3.2 angstrom resolution.
Febs Lett., 594, 2020
|
|
6N5X
 
 | | Crystal structure of the SNX5 PX domain in complex with the CI-MPR (space group P212121 - Form 1) | | Descriptor: | GLYCEROL, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500), Sorting nexin-5,Cation-independent mannose-6-phosphate receptor | | Authors: | Collins, B, Paul, B, Weeratunga, S. | | Deposit date: | 2018-11-22 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Molecular identification of a BAR domain-containing coat complex for endosomal recycling of transmembrane proteins.
Nat.Cell Biol., 21, 2019
|
|
6N5Z
 
 | |
6N5Y
 
 | |
7NS7
 
 | |
8BXA
 
 | | Crystal structure of ribosome binding factor A (RbfA) from S. aureus | | Descriptor: | Ribosome-binding factor A | | Authors: | Fatkhullin, B, Bikmullin, A, Gabdulkhakov, A, Khusainov, I, Validov, S, Usachev, K, Yusupov, M. | | Deposit date: | 2022-12-08 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Yet Another Similarity between Mitochondrial and Bacterial Ribosomal Small Subunit Biogenesis Obtained by Structural Characterization of RbfA from S. aureus.
Int J Mol Sci, 24, 2023
|
|
6SJ5
 
 | | Crystal structure of the uL14-RsfS complex from Staphylococcus aureus | | Descriptor: | 50S ribosomal protein L14, ACETIC ACID, PHOSPHATE ION, ... | | Authors: | Fatkhullin, B, Gabdulkhakov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2019-08-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.26686549 Å) | | Cite: | Mechanism of ribosome shutdown by RsfS in Staphylococcus aureus revealed by integrative structural biology approach.
Nat Commun, 11, 2020
|
|
6SQ8
 
 | | Structure of amide bond synthetase McbA from Marinactinospora thermotolerans | | Descriptor: | 1-ethanoyl-9~{H}-pyrido[3,4-b]indole-3-carboxylic acid, ADENOSINE MONOPHOSPHATE, Fatty acid CoA ligase | | Authors: | Rowlinson, B, Petchey, M, Grogan, G. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Biocatalytic Synthesis of Moclobemide Using the Amide Bond Synthetase McbA Coupled with an ATP Recycling System.
Acs Catalysis, 10, 2020
|
|
6H1B
 
 | | Structure of amide bond synthetase Mcba K483A mutant from Marinactinospora thermotolerans | | Descriptor: | 1-ethanoyl-9~{H}-pyrido[3,4-b]indole-3-carboxylic acid, ADENOSINE MONOPHOSPHATE, Fatty acid CoA ligase | | Authors: | Rowlinson, B, Petchey, M, Cuetos, A, Frese, A, Dannevald, S, Grogan, G. | | Deposit date: | 2018-07-11 | | Release date: | 2018-09-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Broad Aryl Acid Specificity of the Amide Bond Synthetase McbA Suggests Potential for the Biocatalytic Synthesis of Amides.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5TGJ
 
 | |
5TGI
 
 | |
5TGH
 
 | |
1SDU
 
 | | Crystal structures of HIV protease V82A and L90M mutants reveal changes in indinavir binding site. | | Descriptor: | ACETATE ION, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, SULFATE ION, ... | | Authors: | Mahalingam, B, Wang, Y.-F, Boross, P.I, Tozser, J, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2004-02-14 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structures of HIV protease V82A and L90M
mutants reveal changes in the indinavir-binding site
Eur.J.Biochem., 271, 2004
|
|
1SDT
 
 | | Crystal structures of HIV protease V82A and L90M mutants reveal changes in indinavir binding site. | | Descriptor: | CHLORIDE ION, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, protease RETROPEPSIN | | Authors: | Mahalingam, B, Wang, Y.-F, Boross, P.I, Tozser, J, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2004-02-14 | | Release date: | 2004-05-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of HIV protease V82A and L90M
mutants reveal changes in the indinavir-binding site
Eur.J.Biochem., 271, 2004
|
|
1SDV
 
 | | Crystal structures of HIV protease V82A and L90M mutants reveal changes in indinavir binding site. | | Descriptor: | CHLORIDE ION, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, protease RETROPEPSIN | | Authors: | Mahalingam, B, Wang, Y.-F, Boross, P.I, Tozser, J, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2004-02-14 | | Release date: | 2004-05-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of HIV protease V82A and L90M
mutants reveal changes in the indinavir-binding site
Eur.J.Biochem., 271, 2004
|
|
3Q3G
 
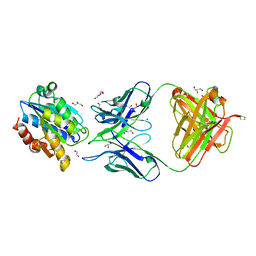 | | Crystal Structure of A-domain in complex with antibody | | Descriptor: | 1,2-ETHANEDIOL, Antibody Heavy chain, Antibody Light Chain, ... | | Authors: | Mahalingam, B, Xiong, J.P, Arnaout, M.A. | | Deposit date: | 2010-12-21 | | Release date: | 2011-11-30 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Stable Coordination of the Inhibitory Ca2+ Ion at the Metal Ion-Dependent Adhesion Site in Integrin CD11b/CD18 by an Antibody-Derived Ligand Aspartate: Implications for Integrin Regulation and Structure-Based Drug Design.
J.Immunol., 187, 2011
|
|
1FG8
 
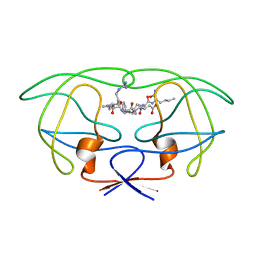 | | STRUCTURAL IMPLICATIONS OF DRUG RESISTANT MUTANTS OF HIV-1 PROTEASE: HIGH RESOLUTION CRYSTAL STRUCTURES OF THE MUTANT PROTEASE/SUBSTRATE ANALOG COMPLEXES | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-07-25 | | Release date: | 2001-06-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes.
Proteins, 43, 2001
|
|
1FFI
 
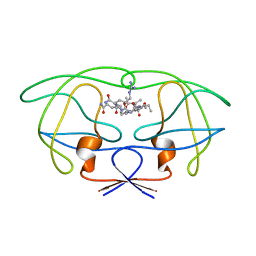 | | STRUCTURAL IMPLICATIONS OF DRUG RESISTANT MUTANTS OF HIV-1 PROTEASE: HIGH RESOLUTION CRYSTAL STRUCTURES OF THE MUTANT PROTEASE/SUBSTRATE ANALOG COMPLEXES | | Descriptor: | N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-07-25 | | Release date: | 2001-06-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes.
Proteins, 43, 2001
|
|
1FGC
 
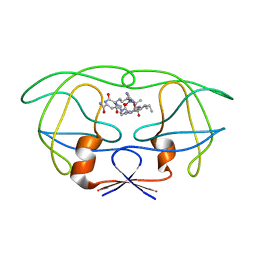 | | STRUCTURAL IMPLICATIONS OF DRUG RESISTANT MUTANTS OF HIV-1 PROTEASE: HIGH RESOLUTION CRYSTAL STRUCTURES OF THE MUTANT PROTEASE/SUBSTRATE ANALOG COMPLEXES | | Descriptor: | N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-07-28 | | Release date: | 2001-06-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes.
Proteins, 43, 2001
|
|
1FFF
 
 | | STRUCTURAL IMPLICATIONS OF DRUG RESISTANT MUTANTS OF HIV-1 PROTEASE : HIGH RESOLUTION CRYSTAL STRUCTURES OF THE MUTANT PROTEASE/SUBSTRATE ANALOG COMPLEXES. | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-07-25 | | Release date: | 2001-06-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes.
Proteins, 43, 2001
|
|
1FEJ
 
 | | STRUCTURAL IMPLICATIONS OF DRUG RESISTANT MUTANTS OF HIV-1 PROTEASE: HIGH RESOLUTION CRYSTAL STRUCTURES OF THE MUTANT PROTEASE/SUBSTRATE ANALOG COMPLEXES | | Descriptor: | N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-07-21 | | Release date: | 2001-06-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes.
Proteins, 43, 2001
|
|
1FG6
 
 | | STRUCTURAL IMPLICATIONS OF DRUG RESISTANT MUTANTS OF HIV-1 PROTEASE: HIGH RESOLUTION CRYSTAL STRUCTURES OF THE MUTANT PROTEASE/SUBSTRATE ANALOG COMPLEXES | | Descriptor: | N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-07-25 | | Release date: | 2001-06-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes.
Proteins, 43, 2001
|
|
1FF0
 
 | | STRUCTURAL IMPLICATIONS OF DRUG RESISTANT MUTANTS OF HIV-1 PROTEASE: HIGH RESOLUTION CRYSTAL STRUCTURES OF THE MUTANT PROTEASE/SUBSTRATE ANALOG COMPLEXES. | | Descriptor: | N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-07-24 | | Release date: | 2001-06-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes.
Proteins, 43, 2001
|
|
3LH9
 
 | |
3LH8
 
 | |
