5ITJ
 
 | | The structure of histone-like protein | | Descriptor: | AbrB family transcriptional regulator, SULFATE ION, TETRAETHYLENE GLYCOL | | Authors: | Lin, B.L, Chen, C.Y, Huang, C.H, Ko, T.P, Chiang, C.H, Lin, K.F, Chang, Y.C, Lin, P.Y, Tsai, H.H.G, Wang, A.H.J. | | Deposit date: | 2016-03-17 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | The Arginine Pairs and C-Termini of the Sso7c4 from Sulfolobus solfataricus Participate in Binding and Bending DNA.
PLoS ONE, 12, 2017
|
|
5ITM
 
 | | The structure of truncated histone-like protein | | Descriptor: | AbrB family transcriptional regulator | | Authors: | Lin, B.L, Chen, C.Y, Huang, C.H, Ko, T.P, Chiang, C.H, Lin, K.F, Chang, Y.C, Lin, P.Y, Tsai, H.H.G, Wang, A.H.J. | | Deposit date: | 2016-03-17 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Arginine Pairs and C-Termini of the Sso7c4 from Sulfolobus solfataricus Participate in Binding and Bending DNA.
PLoS ONE, 12, 2017
|
|
7M59
 
 | |
7M58
 
 | |
8BXA
 
 | | Crystal structure of ribosome binding factor A (RbfA) from S. aureus | | Descriptor: | Ribosome-binding factor A | | Authors: | Fatkhullin, B, Bikmullin, A, Gabdulkhakov, A, Khusainov, I, Validov, S, Usachev, K, Yusupov, M. | | Deposit date: | 2022-12-08 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Yet Another Similarity between Mitochondrial and Bacterial Ribosomal Small Subunit Biogenesis Obtained by Structural Characterization of RbfA from S. aureus.
Int J Mol Sci, 24, 2023
|
|
1NAF
 
 | |
5WK4
 
 | | Crystal structure of an anti-idiotype VLR | | Descriptor: | MAGNESIUM ION, Variable lymphocyte receptor 39 | | Authors: | Collins, B.C, Nakahara, H, Cooper, M.D, Herrin, B.R, Wilson, I.A. | | Deposit date: | 2017-07-24 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Crystal structure of an anti-idiotype variable lymphocyte receptor.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5W8M
 
 | | Crystal structure of Chaetomium thermophilum Vps29 | | Descriptor: | GLYCEROL, TRIETHYLENE GLYCOL, Vacuolar protein sorting-associated protein 29 | | Authors: | Collins, B.M, Leneva, N. | | Deposit date: | 2017-06-21 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure of the membrane-assembled retromer coat determined by cryo-electron tomography.
Nature, 561, 2018
|
|
1EBK
 
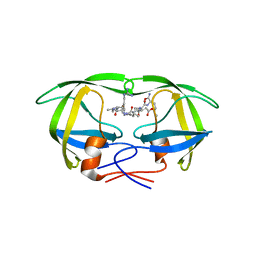 | | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease | | Descriptor: | HIV-1 PROTEASE, N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide | | Authors: | Mahalingam, B, Louis, J.M, Reed, C.C, Adomat, J.M, Krouse, J, Wang, Y.F, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-01-24 | | Release date: | 2000-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease.
Eur.J.Biochem., 263, 1999
|
|
1FG8
 
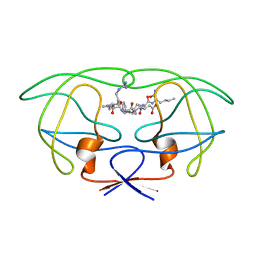 | | STRUCTURAL IMPLICATIONS OF DRUG RESISTANT MUTANTS OF HIV-1 PROTEASE: HIGH RESOLUTION CRYSTAL STRUCTURES OF THE MUTANT PROTEASE/SUBSTRATE ANALOG COMPLEXES | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-07-25 | | Release date: | 2001-06-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes.
Proteins, 43, 2001
|
|
1FFF
 
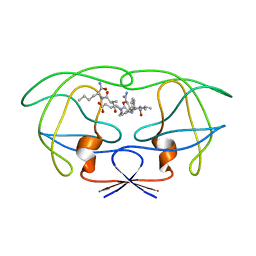 | | STRUCTURAL IMPLICATIONS OF DRUG RESISTANT MUTANTS OF HIV-1 PROTEASE : HIGH RESOLUTION CRYSTAL STRUCTURES OF THE MUTANT PROTEASE/SUBSTRATE ANALOG COMPLEXES. | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-07-25 | | Release date: | 2001-06-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes.
Proteins, 43, 2001
|
|
1FEJ
 
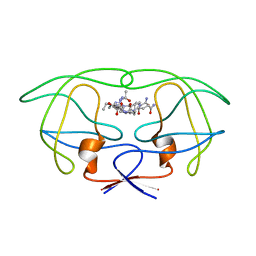 | | STRUCTURAL IMPLICATIONS OF DRUG RESISTANT MUTANTS OF HIV-1 PROTEASE: HIGH RESOLUTION CRYSTAL STRUCTURES OF THE MUTANT PROTEASE/SUBSTRATE ANALOG COMPLEXES | | Descriptor: | N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-07-21 | | Release date: | 2001-06-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes.
Proteins, 43, 2001
|
|
1FG6
 
 | | STRUCTURAL IMPLICATIONS OF DRUG RESISTANT MUTANTS OF HIV-1 PROTEASE: HIGH RESOLUTION CRYSTAL STRUCTURES OF THE MUTANT PROTEASE/SUBSTRATE ANALOG COMPLEXES | | Descriptor: | N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-07-25 | | Release date: | 2001-06-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes.
Proteins, 43, 2001
|
|
7JFY
 
 | | GAS41 YEATS domain in complex with 5 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, N-(5-{3-[(2S)-1,3-thiazolidin-2-yl]azetidine-1-carbonyl}thiophen-2-yl)-L-prolinamide, ... | | Authors: | Linhares, B.M, Listunov, D, Winkler, A, Grembecka, J, Cierpicki, T. | | Deposit date: | 2020-07-17 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.100557 Å) | | Cite: | GAS41 YEATS domain in complex with 5
To Be Published
|
|
1FF0
 
 | | STRUCTURAL IMPLICATIONS OF DRUG RESISTANT MUTANTS OF HIV-1 PROTEASE: HIGH RESOLUTION CRYSTAL STRUCTURES OF THE MUTANT PROTEASE/SUBSTRATE ANALOG COMPLEXES. | | Descriptor: | N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-07-24 | | Release date: | 2001-06-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes.
Proteins, 43, 2001
|
|
6DAQ
 
 | | PhdJ bound to substrate intermediate | | Descriptor: | PhdJ | | Authors: | Medellin, B.P, LeVieux, J.A, Zhang, Y.J, Whitman, C.P. | | Deposit date: | 2018-05-01 | | Release date: | 2019-04-10 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of the Hydratase-Aldolases, NahE and PhdJ: Implications for the Specificity, Catalysis, and N-Acetylneuraminate Lyase Subgroup of the Aldolase Superfamily.
Biochemistry, 57, 2018
|
|
1DW6
 
 | | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease | | Descriptor: | HIV-1 PROTEASE, N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide | | Authors: | Mahalingam, B, Louis, J.M, Reed, C.C, Adomat, J.M, Krouse, J, Wang, Y.F, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-01-24 | | Release date: | 2000-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease.
Eur.J.Biochem., 263, 1999
|
|
6DAN
 
 | | PhdJ WT 2 Angstroms resolution | | Descriptor: | CHLORIDE ION, PhdJ | | Authors: | Medellin, B.P, LeVieux, J.A, Zhang, Y.J, Whitman, C.P. | | Deposit date: | 2018-05-01 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Structural Characterization of the Hydratase-Aldolases, NahE and PhdJ: Implications for the Specificity, Catalysis, and N-Acetylneuraminate Lyase Subgroup of the Aldolase Superfamily.
Biochemistry, 57, 2018
|
|
1FFI
 
 | | STRUCTURAL IMPLICATIONS OF DRUG RESISTANT MUTANTS OF HIV-1 PROTEASE: HIGH RESOLUTION CRYSTAL STRUCTURES OF THE MUTANT PROTEASE/SUBSTRATE ANALOG COMPLEXES | | Descriptor: | N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-07-25 | | Release date: | 2001-06-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes.
Proteins, 43, 2001
|
|
7T58
 
 | | Crystal structure of Miz1 BTB domain | | Descriptor: | DIMETHYL SULFOXIDE, Zinc finger and BTB domain-containing protein 17 | | Authors: | Linhares, B.M, Cierpicki, T. | | Deposit date: | 2021-12-11 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05333114 Å) | | Cite: | Prediction of BTB domain ligandability guided by protein dynamics
To Be Published
|
|
1FGC
 
 | | STRUCTURAL IMPLICATIONS OF DRUG RESISTANT MUTANTS OF HIV-1 PROTEASE: HIGH RESOLUTION CRYSTAL STRUCTURES OF THE MUTANT PROTEASE/SUBSTRATE ANALOG COMPLEXES | | Descriptor: | N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-07-28 | | Release date: | 2001-06-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes.
Proteins, 43, 2001
|
|
6DAO
 
 | | NahE WT selenomethionine | | Descriptor: | Trans-O-hydroxybenzylidenepyruvate hydratase-aldolase | | Authors: | Medellin, B.P, LeVieux, J.A, Zhang, Y.J, Whitman, C.P. | | Deposit date: | 2018-05-01 | | Release date: | 2019-05-08 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.939 Å) | | Cite: | Structural Characterization of the Hydratase-Aldolases, NahE and PhdJ: Implications for the Specificity, Catalysis, and N-Acetylneuraminate Lyase Subgroup of the Aldolase Superfamily.
Biochemistry, 57, 2018
|
|
2EVW
 
 | |
1DAZ
 
 | | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease | | Descriptor: | HIV-1 PROTEASE (RETROPEPSIN), N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide | | Authors: | Mahalingam, B, Louis, J.M, Reed, C.C, Adomat, J.M, Krouse, J, Wang, Y.F, Harrison, R.W, Weber, I.T. | | Deposit date: | 1999-11-01 | | Release date: | 2000-05-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease.
Eur.J.Biochem., 263, 1999
|
|
6MD5
 
 | |
