9B5R
 
 | |
9B5L
 
 | |
9B5I
 
 | |
9B5Q
 
 | |
9B5H
 
 | |
9B5S
 
 | |
9B5X
 
 | |
9B5M
 
 | |
9B5V
 
 | |
9B58
 
 | |
9B5D
 
 | |
9B55
 
 | |
9B59
 
 | |
9B57
 
 | |
9B5C
 
 | |
9B5A
 
 | |
9B56
 
 | |
9B5B
 
 | |
2HVS
 
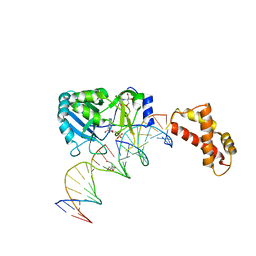 | | Structure of T4 RNA Ligase 2 with Nicked 5'-Adenylated nucleic acid duplex containing a 2'-deoxyribonucleotide at the nick | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*AP*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*TP*TP*G)-3', 5'-D(*CP*AP*AP*TP*TP*GP*CP*GP*AP*C)-R(P*(OMC)P*C)-3', ... | | Authors: | Nandakumar, J, Lima, C.D. | | Deposit date: | 2006-07-30 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA Ligase Structures Reveal the Basis for RNA Specificity and Conformational Changes that Drive Ligation Forward.
Cell(Cambridge,Mass.), 127, 2006
|
|
2HVR
 
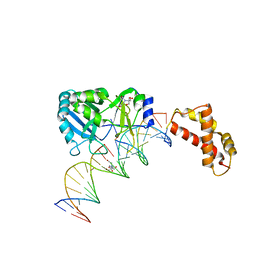 | | Structure of T4 RNA Ligase 2 with Nicked 5'-Adenylated nucleic acid duplex containing a 3'-deoxyribonucleotide at the nick | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*AP*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*TP*TP*G)-3', 5'-D(*CP*AP*AP*TP*TP*GP*CP*GP*AP*C)-R(P*(OMC)P*C)-3', ... | | Authors: | Nandakumar, J, Lima, C.D. | | Deposit date: | 2006-07-30 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | RNA Ligase Structures Reveal the Basis for RNA Specificity and Conformational Changes that Drive Ligation Forward.
Cell(Cambridge,Mass.), 127, 2006
|
|
3SAG
 
 | |
3SAH
 
 | |
3SAF
 
 | |
4II2
 
 | | Crystal structure of Ubiquitin activating enzyme 1 (Uba1) in complex with the Ub E2 Ubc4, ubiquitin, and ATP/Mg | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Olsen, S.K, Lima, C.D. | | Deposit date: | 2012-12-19 | | Release date: | 2013-02-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a ubiquitin E1-E2 complex: insights to E1-E2 thioester transfer.
Mol.Cell, 49, 2013
|
|
3EF0
 
 | |
