4I52
 
 | | scMenB im complex with 1-hydroxy-2-naphthoyl-CoA | | Descriptor: | 1-hydroxy-2-naphthoyl-CoA, CHLORIDE ION, Naphthoate synthase | | Authors: | Song, H.G, Sun, Y.R, Li, J, Li, Y, Jiang, M, Zhou, J.H, Guo, Z.H. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of the induced-fit mechanism of 1,4-dihydroxy-2-naphthoyl coenzyme a synthase from the crotonase fold superfamily
Plos One, 8, 2013
|
|
4I4Z
 
 | | Synechocystis sp. PCC 6803 1,4-dihydroxy-2-naphthoyl-coenzyme A synthase (MenB) in complex with salicylyl-CoA | | Descriptor: | BICARBONATE ION, MALONATE ION, Naphthoate synthase, ... | | Authors: | Song, H.G, Sun, Y.R, Li, J, Li, Y, Jiang, M, Zhou, J.H, Guo, Z.H. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the induced-fit mechanism of 1,4-dihydroxy-2-naphthoyl coenzyme a synthase from the crotonase fold superfamily
Plos One, 8, 2013
|
|
6CMX
 
 | | Human Teneurin 2 extra-cellular region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Teneurin-2, alpha-D-mannopyranose, ... | | Authors: | Shalev-Benami, M, Li, J, Sudhof, T, Skiniotis, G, Arac, D. | | Deposit date: | 2018-03-06 | | Release date: | 2018-07-25 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis for Teneurin Function in Circuit-Wiring: A Toxin Motif at the Synapse.
Cell, 173, 2018
|
|
5GGZ
 
 | | Crystal structure of novel inhibitor bound with Hsp90 | | Descriptor: | Heat shock protein HSP 90-alpha, [2,4-bis(oxidanyl)-5-propan-2-yl-phenyl]-(2-ethoxy-7,8-dihydro-5~{H}-pyrido[4,3-d]pyrimidin-6-yl)methanone | | Authors: | Chen, T.T, Li, J, Xu, Y.C. | | Deposit date: | 2016-06-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Novel Tetrahydropyrido[4,3-d]pyrimidines as Potent Inhibitors of Chaperone Heat Shock Protein 90
J. Med. Chem., 59, 2016
|
|
3SR9
 
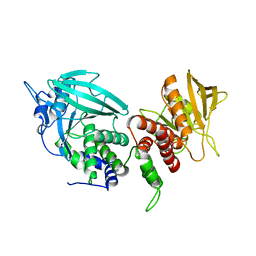 | | Crystal structure of mouse PTPsigma | | Descriptor: | Receptor-type tyrosine-protein phosphatase S | | Authors: | Wang, J, Hou, L, Li, J, Ding, J. | | Deposit date: | 2011-07-07 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the homology and differences between mouse protein tyrosine phosphatase-sigma and human protein tyrosine phosphatase-sigma
Acta Biochim.Biophys.Sin., 43, 2011
|
|
7FB5
 
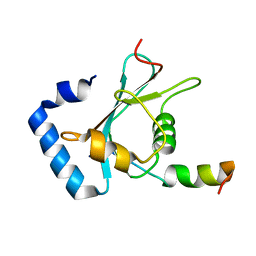 | | Crystal structure of FAM134B/GABARAP complex | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein, Reticulophagy regulator 1 | | Authors: | Zhao, J, Li, J. | | Deposit date: | 2021-07-08 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | The crystal structure of the FAM134B-GABARAP complex provides mechanistic insights into the selective binding of FAM134 to the GABARAP subfamily.
Febs Open Bio, 12, 2022
|
|
8FV5
 
 | | Representation of 16-mer phiPA3 PhuN Lattice, p2 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein, phiPA3 PhuN | | Authors: | Nieweglowska, E.S, Brilot, A.F, Mendez-Moran, M, Kokontis, C, Baek, M, Li, J, Cheng, Y, Baker, D, Bondy-Denomy, J, Agard, D.A. | | Deposit date: | 2023-01-18 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.21 Å) | | Cite: | The phi PA3 phage nucleus is enclosed by a self-assembling 2D crystalline lattice.
Nat Commun, 14, 2023
|
|
5Z2N
 
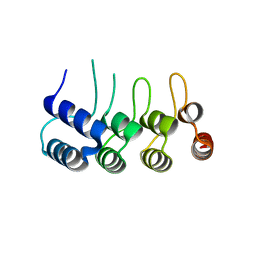 | | Structure of Orp1L N-terminal Domain | | Descriptor: | Oxysterol-binding protein-related protein 1 | | Authors: | Ma, X.L, Liu, K, Li, J, Li, H.H, Li, J, Yang, C.L, Liang, H.H. | | Deposit date: | 2018-01-03 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | A non-canonical GTPase interaction enables ORP1L-Rab7-RILP complex formation and late endosome positioning.
J. Biol. Chem., 293, 2018
|
|
5Z96
 
 | | Structure of the mouse TRPC4 ion channel | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, CHOLESTEROL HEMISUCCINATE, SODIUM ION, ... | | Authors: | Duan, J, Li, Z, Li, J, Zhang, J. | | Deposit date: | 2018-02-02 | | Release date: | 2018-04-18 | | Last modified: | 2018-08-29 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure of the mouse TRPC4 ion channel.
Nat Commun, 9, 2018
|
|
5ZZ9
 
 | | Crystal structure of Homer2 EVH1/Drebrin PPXXF complex | | Descriptor: | Homer protein homolog 2, Peptide from Drebrin | | Authors: | Li, Z, Liu, H, Li, J, Liu, W, Zhang, M. | | Deposit date: | 2018-05-31 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Homer Tetramer Promotes Actin Bundling Activity of Drebrin.
Structure, 27, 2019
|
|
1W99
 
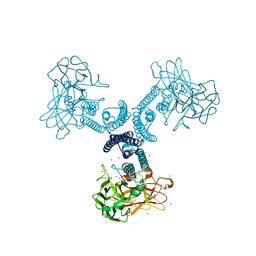 | |
8IG7
 
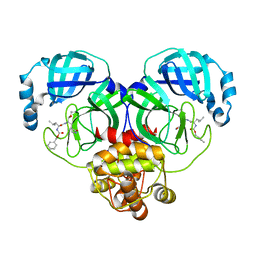 | |
8IG4
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with GC376 | | Descriptor: | Non-structural protein 11, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Zhou, X.L, Zhang, J, Li, J. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Coronaviral Main Proteases Inhibition by the 3CLpro Inhibitor GC376.
J.Mol.Biol., 436, 2024
|
|
8IG5
 
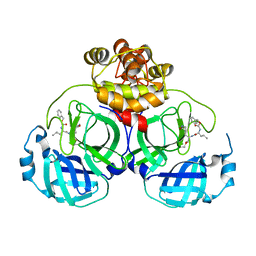 | | Crystal structure of SARS main protease in complex with GC376 | | Descriptor: | 3C-like proteinase nsp5, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Lin, C, Zhang, J, Li, J. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural Basis for Coronaviral Main Proteases Inhibition by the 3CLpro Inhibitor GC376.
J.Mol.Biol., 436, 2024
|
|
8IG6
 
 | | Crystal structure of MERS main protease in complex with GC376 | | Descriptor: | N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide, ORF1a | | Authors: | Lin, C, Zhang, J, Li, J. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural Basis for Coronaviral Main Proteases Inhibition by the 3CLpro Inhibitor GC376.
J.Mol.Biol., 436, 2024
|
|
8IG8
 
 | |
8IG9
 
 | |
5WQT
 
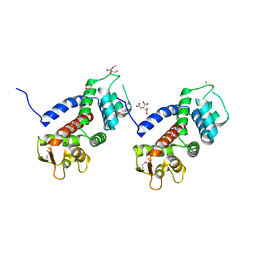 | | Structure of a protein involved in pyroptosis | | Descriptor: | CITRIC ACID, GLYCEROL, Gasdermin-D | | Authors: | Kuang, S, Li, J. | | Deposit date: | 2016-11-28 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure insight of GSDMD reveals the basis of GSDMD autoinhibition in cell pyroptosis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1BWP
 
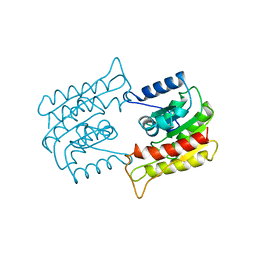 | | PROBING THE SUBSTRATE SPECIFICITY OF THE INTRACELLULAR BRAIN PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Descriptor: | PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Authors: | Ho, Y.S, Sheffield, P.J, Masuyama, J, Arai, H, Li, J, Aoki, J, Inoue, K, Derewenda, U, Derewenda, Z. | | Deposit date: | 1998-09-27 | | Release date: | 1999-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the Substrate Specificity of the Intracellular Brain Platelet-Activating Factor Acetylhydrolase
Protein Eng., 12, 1999
|
|
1BWR
 
 | | PROBING THE SUBSTRATE SPECIFICITY OF THE INTRACELLULAR BRAIN PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Descriptor: | PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Authors: | Ho, Y.S, Sheffield, P.J, Masuyama, J, Arai, H, Li, J, Aoki, J, Inoue, K, Derewenda, U, Derewenda, Z. | | Deposit date: | 1998-09-27 | | Release date: | 1999-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the Substrate Specificity of the Intracellular Brain Platelet-Activating Factor Acetylhydrolase
Protein Eng., 12, 1999
|
|
5XF7
 
 | |
4FWV
 
 | | Crystal structure of the N-terminal domain of the Lon-like protease MtaLonC | | Descriptor: | SULFATE ION, TTC1975 peptidase | | Authors: | Chang, C.I, Li, J.K, Kuo, C.I, Huang, K.F. | | Deposit date: | 2012-07-02 | | Release date: | 2013-06-26 | | Last modified: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The N-terminal substrate-recognition domain of a LonC protease exhibits structural and functional similarity to cytosolic chaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7C29
 
 | | Esterase CrmE10 mutant-D178A | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Li, Z, Li, J. | | Deposit date: | 2020-05-07 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure-guided protein engineering increases enzymatic activities of the SGNH family esterases.
Biotechnol Biofuels, 13, 2020
|
|
8H93
 
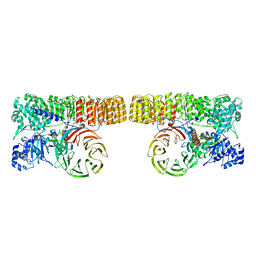 | | Structure of dimeric mouse SCMC core complex | | Descriptor: | NACHT, LRR and PYD domains-containing protein 5, Oocyte-expressed protein homolog, ... | | Authors: | Chi, P, Ou, G, Han, Z, Li, J, Deng, D. | | Deposit date: | 2022-10-24 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis of the subcortical maternal complex and its implications in reproductive disorders.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H94
 
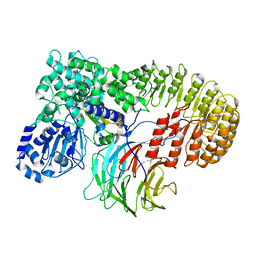 | | Structure of mouse SCMC bound with KH domain of FILIA | | Descriptor: | NACHT, LRR and PYD domains-containing protein 5, Oocyte-expressed protein homolog, ... | | Authors: | Chi, P, Ou, G, Han, Z, Li, J, Deng, D. | | Deposit date: | 2022-10-24 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of the subcortical maternal complex and its implications in reproductive disorders.
Nat.Struct.Mol.Biol., 31, 2024
|
|
