5D8E
 
 | |
3N54
 
 | | Crystal Structure of the GerBC protein | | Descriptor: | CHLORIDE ION, SULFATE ION, Spore germination protein B3 | | Authors: | Li, Y, Setlow, B, Setlow, P, Hao, B. | | Deposit date: | 2010-05-24 | | Release date: | 2010-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the GerBC Component of a Bacillus subtilis Spore Germinant Receptor.
J.Mol.Biol., 402, 2010
|
|
5DO2
 
 | | Complex structure of MERS-RBD bound with 4C2 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4C2 heavy chain, 4C2 light chain, ... | | Authors: | Li, Y, Wan, Y, Liu, P, Zhao, J, Lu, G, Qi, J, Wang, Q, Lu, X, Wu, Y, Liu, W, Yuen, K.Y, Perlman, S, Gao, G.F, Yan, J. | | Deposit date: | 2015-09-10 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.409 Å) | | Cite: | A humanized neutralizing antibody against MERS-CoV targeting the receptor-binding domain of the spike protein.
Cell Res., 25, 2015
|
|
3BW4
 
 | |
2HB0
 
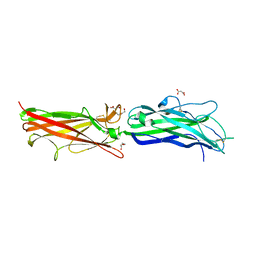 | | Crystal Structure of CfaE, the Adhesive Subunit of CFA/I Fimbria of Enterotoxigenic Escherichia coli | | Descriptor: | CFA/I fimbrial subunit E, DI(HYDROXYETHYL)ETHER, MALONATE ION | | Authors: | Li, Y.F, Xia, D, Poole, S, Rasulova, F, Savarino, S.J. | | Deposit date: | 2006-06-13 | | Release date: | 2007-06-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A receptor-binding site as revealed by the crystal structure of CfaE, the colonization factor antigen I fimbrial adhesin of enterotoxigenic Escherichia coli.
J.Biol.Chem., 282, 2007
|
|
4P5A
 
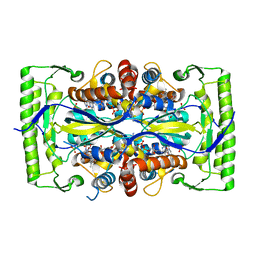 | | Crystal structure of a UMP/dUMP methylase PolB from Streptomyces cacaoi bound with 5-Br UMP | | Descriptor: | 5-BROMO-URIDINE-5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, Thymidylate synthase ThyX | | Authors: | Li, Y, Chen, W, Li, J, Xia, Z, Deng, Z, Zhou, J. | | Deposit date: | 2014-03-15 | | Release date: | 2015-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of a UMP/dUMP methylase PolB from Streptomyces cacaoi with 5-Br UMP
To Be Published
|
|
4P5B
 
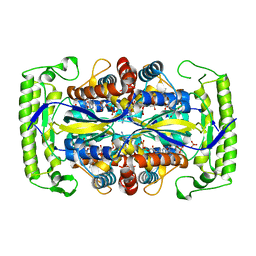 | | Crystal structure of a UMP/dUMP methylase PolB from Streptomyces cacaoi bound with 5-Br dUMP | | Descriptor: | 5-BROMO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Li, Y, Chen, W, Li, J, Xia, Z, Deng, Z, Zhou, J. | | Deposit date: | 2014-03-15 | | Release date: | 2015-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.274 Å) | | Cite: | Crystal structure of a UMP/dUMP methylase PolB form Streptomyces cacaoi bound with 5-Br dUMP
To Be Published
|
|
5ZN9
 
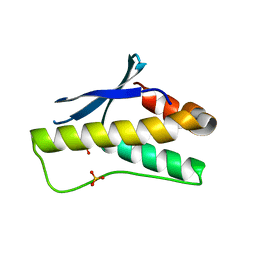 | | Crystal structure of PX domain | | Descriptor: | SULFATE ION, Sorting nexin-27 | | Authors: | Li, Y, Zhu, Z, Li, F, Liao, S, Xu, C. | | Deposit date: | 2018-04-08 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.776 Å) | | Cite: | Crystal structure of PX domain
To Be Published
|
|
3JBB
 
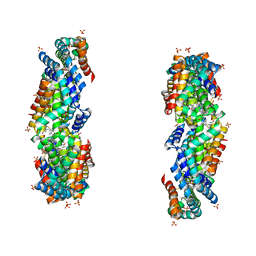 | | Characterization of red-shifted phycobiliprotein complexes isolated from the chlorophyll f-containing cyanobacterium Halomicronema hongdechloris | | Descriptor: | PHYCOCYANOBILIN, SULFATE ION, allophycocyanin beta chain, ... | | Authors: | Li, Y, Lin, Y, Garvey, C, Birch, D, Corkery, R.W, Loughlin, P.C, Scheer, H, Willows, R.D, Chen, M. | | Deposit date: | 2015-08-26 | | Release date: | 2015-11-11 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (26 Å) | | Cite: | Characterization of red-shifted phycobilisomes isolated from the chlorophyll f-containing cyanobacterium Halomicronema hongdechloris.
Biochim.Biophys.Acta, 1857, 2015
|
|
3DXF
 
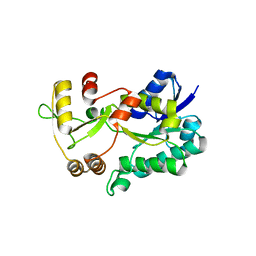 | | Crystal structure of the HSCARG R37A mutant | | Descriptor: | NmrA-like family domain-containing protein 1 | | Authors: | Li, Y, Meng, G, Dai, X, Luo, M, Zheng, X. | | Deposit date: | 2008-07-24 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | NADPH is an allosteric regulator of HSCARG
J.Mol.Biol., 387, 2009
|
|
3E5M
 
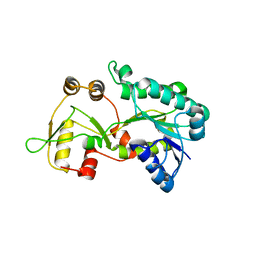 | | Crystal structure of the HSCARG Y81A mutant | | Descriptor: | NmrA-like family domain-containing protein 1 | | Authors: | Li, Y, Meng, G, Dai, X, Luo, M, Zheng, X. | | Deposit date: | 2008-08-14 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | NADPH is an allosteric regulator of HSCARG
J.Mol.Biol., 387, 2009
|
|
7X8F
 
 | |
7X8B
 
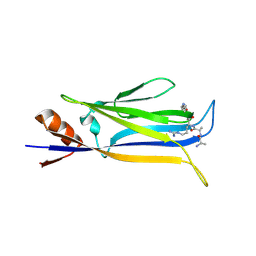 | |
7X8G
 
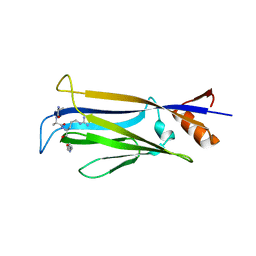 | |
1SKR
 
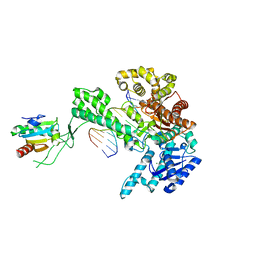 | | T7 DNA Polymerase Complexed To DNA Primer/Template and ddATP | | Descriptor: | 2',3'-DIDEOXYADENOSINE-5'-TRIPHOSPHATE, 5'-D(*CP*CP*CP*TP*TP*TP*GP*GP*CP*AP*CP*TP*GP*GP*CP*CP*GP*TP*CP*GP*TP*TP*TP*TP*CP*G)-3', 5'-D(*CP*GP*AP*AP*AP*AP*CP*GP*AP*C*GP*GP*CP*CP*AP*GP*TP*GP*CP*CP*AP*(2DA))-3', ... | | Authors: | Li, Y, Dutta, S, Doublie, S, Bdour, H.M, Taylor, J.S, Ellenberger, T. | | Deposit date: | 2004-03-05 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nucleotide insertion opposite a cis-syn thymine dimer by a replicative DNA polymerase from bacteriophage T7.
Nat.Struct.Mol.Biol., 11, 2004
|
|
2FJ2
 
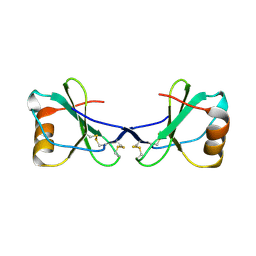 | | Crystal Structure of Viral Macrophage Inflammatory Protein-II | | Descriptor: | Viral macrophage inflammatory protein-II | | Authors: | Li, Y, Liu, D, Cao, R, Kumar, S, Dong, C.Z, wilson, S.R, Gao, Y.G, Huang, Z. | | Deposit date: | 2005-12-30 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of chemically synthesized vMIP-II.
Proteins, 67, 2007
|
|
2FHT
 
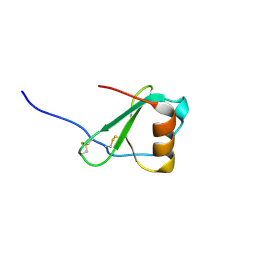 | | Crystal Structure of Viral Macrophage Inflammatory Protein-II | | Descriptor: | Viral macrophage inflammatory protein-II | | Authors: | Li, Y, Liu, D, Cao, R, Kumar, S, Dong, C.Z, wilson, S.R, Gao, Y.G, Huang, Z. | | Deposit date: | 2005-12-27 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of chemically synthesized vMIP-II.
Proteins, 67, 2007
|
|
2M19
 
 | | Solution structure of the Haloferax volcanii HVO 2177 protein | | Descriptor: | Molybdopterin converting factor subunit 1 | | Authors: | Li, Y, Maciejewski, M.W, Martin, J, Jin, K, Zhang, Y, Lu, M, Maupin-Furlow, J.A, Hao, B. | | Deposit date: | 2012-11-21 | | Release date: | 2013-08-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Crystal structure of the ubiquitin-like small archaeal modifier protein 2 from Haloferax volcanii.
Protein Sci., 22, 2013
|
|
1WPA
 
 | |
1XOO
 
 | | NMR structure of G1S mutant of influenza hemagglutinin fusion peptide in DPC micelles at pH 5 | | Descriptor: | Hemagglutinin | | Authors: | Li, Y, Han, X, Lai, A.L, Bushweller, J.H, Cafiso, D.S, Tamm, L.K. | | Deposit date: | 2004-10-06 | | Release date: | 2005-09-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Membrane structures of the hemifusion-inducing fusion peptide mutant G1S and the fusion-blocking mutant G1V of influenza virus hemagglutinin suggest a mechanism for pore opening in membrane fusion.
J.Virol., 79, 2005
|
|
1XAW
 
 | |
1XOP
 
 | | NMR structure of G1V mutant of influenza hemagglutinin fusion peptide in DPC micelles at pH 5 | | Descriptor: | Hemagglutinin | | Authors: | Li, Y, Han, X, Lai, A.L, Bushweller, J.H, Cafiso, D.S, Tamm, L.K. | | Deposit date: | 2004-10-06 | | Release date: | 2005-09-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Membrane structures of the hemifusion-inducing fusion peptide mutant G1S and the fusion-blocking mutant G1V of influenza virus hemagglutinin suggest a mechanism for pore opening in membrane fusion.
J.Virol., 79, 2005
|
|
6A3V
 
 | | Complex structure of human 4-1BB and 4-1BBL | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 9, Tumor necrosis factor receptor superfamily member 9 | | Authors: | Li, Y, Zhang, C, Chai, Y, Qi, J, Tien, P, Gao, S, Gao, G.F. | | Deposit date: | 2018-06-17 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.391 Å) | | Cite: | Limited Cross-Linking of 4-1BB by 4-1BB Ligand and the Agonist Monoclonal Antibody Utomilumab.
Cell Rep, 25, 2018
|
|
1YP0
 
 | | Structure of the steroidogenic factor-1 ligand binding domain bound to phospholipid and a SHP peptide motif | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Nuclear receptor subfamily 0, group B, ... | | Authors: | Li, Y, Choi, M, Cavey, G, Daugherty, J, Suino, K, Kovach, A, Bingham, N, Kliewer, S, Xu, H. | | Deposit date: | 2005-01-28 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic identification and functional characterization of phospholipids as ligands for the orphan nuclear receptor steroidogenic factor-1.
Mol.Cell, 17, 2005
|
|
1ZGY
 
 | | Structural and Biochemical Basis for Selective Repression of the Orphan Nuclear Receptor LRH-1 by SHP | | Descriptor: | 2,4-THIAZOLIDIINEDIONE, 5-[[4-[2-(METHYL-2-PYRIDINYLAMINO)ETHOXY]PHENYL]METHYL]-(9CL), Nuclear receptor subfamily 0, ... | | Authors: | Li, Y, Choi, M, Suino, K, Kovach, A, Daugherty, J, Kliewer, S.A, Xu, H.E. | | Deposit date: | 2005-04-22 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and biochemical basis for selective repression of the orphan nuclear receptor liver receptor homolog 1 by small heterodimer partner.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
