2FQF
 
 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | Descriptor: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
4KMP
 
 | | Structure of XIAP-BIR3 and inhibitor | | Descriptor: | (2S,2'S)-N,N'-[(6,6'-difluoro-1H,1'H-2,2'-biindole-3,3'-diyl)bis{methanediyl[(2R,4S)-4-hydroxypyrrolidine-2,1-diyl][(2S)-1-oxobutane-1,2-diyl]}]bis[2-(methylamino)propanamide], E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Li, X, Wang, J, Condon, S.M, Shi, Y. | | Deposit date: | 2013-05-08 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of XIAP-BIR3 and inhibitor
To be Published
|
|
2FQE
 
 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | Descriptor: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
2FQG
 
 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | Descriptor: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
7CR7
 
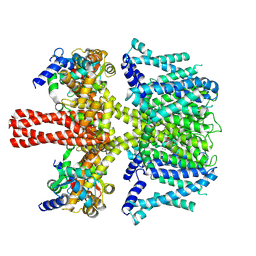 | | human KCNQ2-CaM in complex with retigabine | | Descriptor: | Calmodulin-3, Potassium voltage-gated channel subfamily KQT member 2, ethyl N-[2-azanyl-4-[(4-fluorophenyl)methylamino]phenyl]carbamate | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
7CR0
 
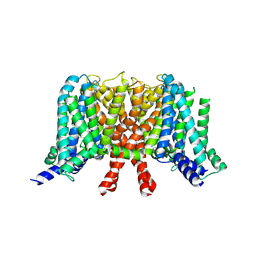 | | human KCNQ2 in apo state | | Descriptor: | Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
7CR3
 
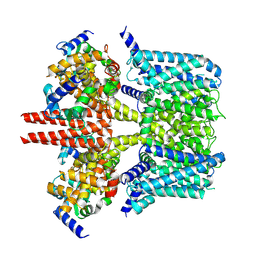 | | human KCNQ2-CaM in apo state | | Descriptor: | Calmodulin-3, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
7CR4
 
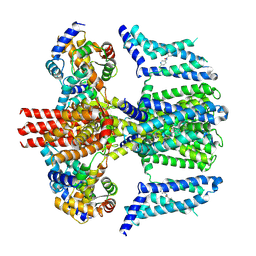 | | human KCNQ2-CaM in complex with ztz240 | | Descriptor: | Calmodulin-3, N-(6-chloranylpyridin-3-yl)-4-fluoranyl-benzamide, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
7CR1
 
 | | human KCNQ2 in complex with ztz240 | | Descriptor: | N-(6-chloranylpyridin-3-yl)-4-fluoranyl-benzamide, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
5YGI
 
 | |
6K32
 
 | | RdRp complex | | Descriptor: | 2'-O-methyladenosine 5'-(dihydrogen phosphate), 7-METHYLGUANOSINE, DIPHOSPHATE, ... | | Authors: | Li, X.W. | | Deposit date: | 2019-05-16 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of RdRps Within a Transcribing dsRNA Virus Provides Insights Into the Mechanisms of RNA Synthesis.
J.Mol.Biol., 432, 2020
|
|
8HPE
 
 | | Crystal structure of Leucine dehydrogenase | | Descriptor: | GLYCEROL, Leucine dehydrogenase, SULFATE ION | | Authors: | Li, X, Song, W. | | Deposit date: | 2022-12-12 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | A Tri-Enzyme Cascade for Efficient Production of L-2-Aminobutyrate from L-Threonine.
Chembiochem, 24, 2023
|
|
7VS7
 
 | | Crystal structure of the ectodomain of OsCERK1 in complex with chitin hexamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin elicitor receptor kinase 1, ... | | Authors: | Li, X. | | Deposit date: | 2021-10-26 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Structural insight into chitin perception by chitin elicitor receptor kinase 1 of Oryza sativa.
J Integr Plant Biol, 65, 2023
|
|
4HYD
 
 | | Structure of a presenilin family intramembrane aspartate protease in C2221 space group | | Descriptor: | Putative uncharacterized protein | | Authors: | Li, X, Dang, S, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2012-11-13 | | Release date: | 2012-12-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of a presenilin family intramembrane aspartate protease
Nature, 493, 2013
|
|
4HYC
 
 | | Structure of a presenilin family intramembrane aspartate protease in P2 space group | | Descriptor: | Putative uncharacterized protein | | Authors: | Li, X, Dang, S, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2012-11-13 | | Release date: | 2012-12-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Structure of a presenilin family intramembrane aspartate protease
Nature, 493, 2013
|
|
8HR6
 
 | |
1CS3
 
 | |
8IET
 
 | |
6KPF
 
 | | Cryo-EM structure of a class A GPCR with G protein complex | | Descriptor: | 7-[(6aR,9R,10aR)-1-Hydroxy-9-(hydroxymethyl)-6,6-dimethyl-6a,7,8,9,10,10a-hexahydro-6H-benzo[c]chromen-3-yl]- 7-methyloctanenitrile, Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, X.T, Hua, T, Wu, L.J, Makriyannis, A, Shen, L, Wang, Y.X, Liu, Z.J. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-12 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-GiComplex Structures.
Cell, 180, 2020
|
|
4JPB
 
 | | The structure of a ternary complex between CheA domains P4 and P5 with CheW and with an unzipped fragment of TM14, a chemoreceptor analog from Thermotoga maritima. | | Descriptor: | Chemotaxis protein CheA, Chemotaxis protein CheW, Methyl-accepting chemotaxis protein | | Authors: | Li, X, Bayas, C, Bilwes, A.M, Crane, B.R. | | Deposit date: | 2013-03-19 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.186 Å) | | Cite: | The 3.2 angstrom resolution structure of a receptor: CheA:CheW signaling complex defines overlapping binding sites and key residue interactions within bacterial chemosensory arrays.
Biochemistry, 52, 2013
|
|
7VBR
 
 | |
6KPC
 
 | | Crystal structure of an agonist bound GPCR | | Descriptor: | 7-[(6aR,9R,10aR)-1-Hydroxy-9-(hydroxymethyl)-6,6-dimethyl-6a,7,8,9,10,10a-hexahydro-6H-benzo[c]chromen-3-yl]- 7-methyloctanenitrile, Cannabinoid receptor 2,Endolysin | | Authors: | Li, X.T, Hua, T, Wu, L.J, Makriyannis, A, Wu, M, Liu, Z.J. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-GiComplex Structures.
Cell, 180, 2020
|
|
4HYG
 
 | | Structure of a presenilin family intramembrane aspartate protease in C222 space group | | Descriptor: | Putative uncharacterized protein | | Authors: | Li, X, Dang, S, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2012-11-13 | | Release date: | 2012-12-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Structure of a presenilin family intramembrane aspartate protease
Nature, 493, 2013
|
|
5X5S
 
 | |
7WJG
 
 | | Streptococcus mutans BusR transcriptional factor | | Descriptor: | CITRIC ACID, Putative transcriptional regulator (GntR family) | | Authors: | Li, X, Liu, N. | | Deposit date: | 2022-01-06 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural study of transcriptional factor BusR
To Be Published
|
|
