5NKK
 
 | | SMG8-SMG9 complex GDP bound | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Li, L, Basquin, J, Conti, E. | | Deposit date: | 2017-03-31 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure of a SMG8-SMG9 complex identifies a G-domain heterodimer in the NMD effector proteins.
RNA, 23, 2017
|
|
4FQA
 
 | |
8E13
 
 | |
8E8I
 
 | |
8EC5
 
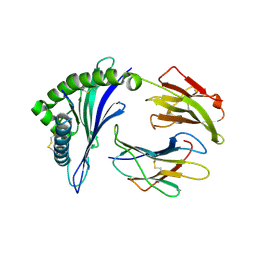 | |
8E2Z
 
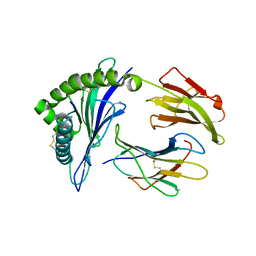 | |
6JCX
 
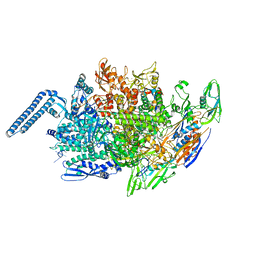 | |
6JCY
 
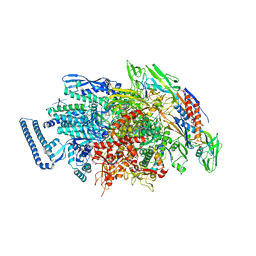 | |
9J1D
 
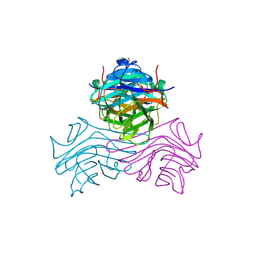 | | Structure of ConA/PHE-Man | | Descriptor: | CALCIUM ION, Concanavalin-A, MANGANESE (II) ION, ... | | Authors: | Li, L, Chen, G. | | Deposit date: | 2024-08-04 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Structure of ConA/PHE-Man
To Be Published
|
|
9J1C
 
 | | Structure of ConA/NAP-Man | | Descriptor: | CALCIUM ION, Concanavalin-A, MANGANESE (II) ION, ... | | Authors: | Li, L, Chen, G. | | Deposit date: | 2024-08-04 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure of ConA/NAP-Man
To Be Published
|
|
9J1F
 
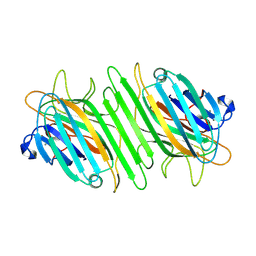 | | Dimeric Structure of ConA/M2P-Man | | Descriptor: | CALCIUM ION, Concanavalin-A, MANGANESE (II) ION, ... | | Authors: | Li, L, Chen, G. | | Deposit date: | 2024-08-04 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Dimeric Structure of ConA/M2P-Man
To Be Published
|
|
9J1A
 
 | | Structure of ConA without ligand | | Descriptor: | CALCIUM ION, Concanavalin-A, GLYCEROL, ... | | Authors: | Li, L, Chen, G. | | Deposit date: | 2024-08-04 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure of ConA without ligand
To Be Published
|
|
9J1B
 
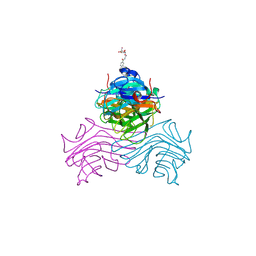 | | Structure of ConA/IMI-Man | | Descriptor: | CALCIUM ION, Concanavalin-A, MANGANESE (II) ION, ... | | Authors: | Li, L, Chen, G. | | Deposit date: | 2024-08-04 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure of ConA/IMI-Man
To Be Published
|
|
8FNZ
 
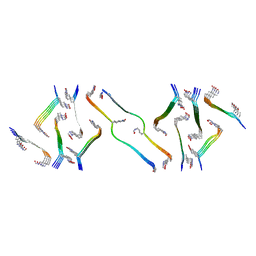 | | Acetylated tau repeat 1 and 2 fragment (AcR1R2) | | Descriptor: | Microtubule-associated protein tau, acetylated repeat 1 and 2 fragment | | Authors: | Li, L, Nguyen, A.B, Mullapudi, V, Joachimiak, L. | | Deposit date: | 2022-12-29 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Disease-associated patterns of acetylation stabilize tau fibril formation.
Structure, 31, 2023
|
|
8WMK
 
 | | Structure of ConA/Man2 | | Descriptor: | 4-[3-(4-carboxyphenyl)phenyl]benzoic acid, CALCIUM ION, Concanavalin-A, ... | | Authors: | Li, L, Chen, G. | | Deposit date: | 2023-10-03 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Rational design of inducing ligands with three-dimensional supramolecular interactions to build protein crystalline frameworks.
To Be Published
|
|
8WMG
 
 | | Structure of ConA/Man3 | | Descriptor: | 4-[3,5-bis(4-carboxyphenyl)phenyl]benzoic acid, CALCIUM ION, Concanavalin-A, ... | | Authors: | Li, L, Chen, G. | | Deposit date: | 2023-10-03 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Rational design of inducing ligands with three-dimensional supramolecular interactions to build protein crystalline frameworks.
To Be Published
|
|
3ZIU
 
 | | Crystal structure of Mycoplasma mobile Leucyl-tRNA Synthetase with Leu-AMS in the active site | | Descriptor: | 5'-O-(L-leucylsulfamoyl)adenosine, GLYCEROL, LEUCYL-TRNA SYNTHETASE | | Authors: | Li, L, Palencia, A, Lukk, T, Li, Z, Luthey-Schulten, Z.A, Cusack, S, Martinis, S.A, Boniecki, M.T. | | Deposit date: | 2013-01-10 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Leucyl-tRNA Synthetase Editing Domain Functions as a Molecular Rheostat to Control Codon Ambiguity in Mycoplasma Pathogens.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3B9O
 
 | | long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN | | Descriptor: | Alkane monooxygenase, FLAVIN MONONUCLEOTIDE | | Authors: | Li, L, Yang, W, Xu, F, Bartlam, M, Rao, Z. | | Deposit date: | 2007-11-06 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN: unveiling the long-chain alkane hydroxylase
J.Mol.Biol., 376, 2008
|
|
3B9N
 
 | | Crystal structure of long-chain alkane monooxygenase (LadA) | | Descriptor: | Alkane monooxygenase | | Authors: | Li, L, Yang, W, Xu, F, Bartlam, M, Rao, Z. | | Deposit date: | 2007-11-06 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN: unveiling the long-chain alkane hydroxylase
J.Mol.Biol., 376, 2008
|
|
5NKM
 
 | | SMG8-SMG9 complex | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, L, Basquin, J, Conti, E. | | Deposit date: | 2017-03-31 | | Release date: | 2017-04-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Structure of a SMG8-SMG9 complex identifies a G-domain heterodimer in the NMD effector proteins.
RNA, 23, 2017
|
|
7SH0
 
 | | CRYSTAL STRUCTURE OF ENDOPLASMIC RETICULUM AMINOPEPTIDASE 2 (ERAP2) COMPLEX WITH A HIGHLY SELECTIVE AND POTENT SMALL MOLECULE | | Descriptor: | (2S)-N-hydroxy-3-(4-methoxyphenyl)-2-[4-({[5-(pyridin-2-yl)thiophene-2-sulfonyl]amino}methyl)-1H-1,2,3-triazol-1-yl]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Li, L, Bouvier, M. | | Deposit date: | 2021-10-07 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of the First Selective Nanomolar Inhibitors of ERAP2 by Kinetic Target-Guided Synthesis.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
1QVO
 
 | | STRUCTURES OF HLA-A*1101 IN COMPLEX WITH IMMUNODOMINANT NONAMER AND DECAMER HIV-1 EPITOPES CLEARLY REVEAL THE PRESENCE OF A MIDDLE ANCHOR RESIDUE | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-11 alpha chain, ... | | Authors: | Li, L, McNicholl, J.M, Bouvier, M. | | Deposit date: | 2003-08-28 | | Release date: | 2004-06-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structures of HLA-A*1101 complexed with immunodominant nonamer and decamer HIV-1 epitopes clearly reveal the presence of a middle, secondary anchor residue.
J.Immunol., 172, 2004
|
|
2PQ6
 
 | |
7N8T
 
 | | Crystal Structure of AMP-bound Human JNK2 | | Descriptor: | ADENOSINE MONOPHOSPHATE, HEXAETHYLENE GLYCOL, Mitogen-activated protein kinase 9 | | Authors: | Li, L, Gurbani, D, Westover, K.D. | | Deposit date: | 2021-06-15 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Development of a Covalent Inhibitor of c-Jun N-Terminal Protein Kinase (JNK) 2/3 with Selectivity over JNK1.
J.Med.Chem., 66, 2023
|
|
6P27
 
 | | Structure of a nested set of N-terminally extended MHC I-peptides provides novel insights into antigen processing and presentation | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-8 alpha chain, ... | | Authors: | Li, L, Batliwala, M, Bouvier, M. | | Deposit date: | 2019-05-21 | | Release date: | 2019-10-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.593 Å) | | Cite: | ERAP1 enzyme-mediated trimming and structural analyses of MHC I-bound precursor peptides yield novel insights into antigen processing and presentation.
J.Biol.Chem., 294, 2019
|
|
