4OHU
 
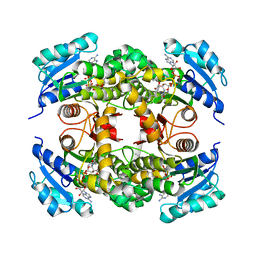 | | Crystal structure of Mycobacterium tuberculosis InhA in complex with inhibitor PT92 | | Descriptor: | 2-(2-bromophenoxy)-5-hexylphenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Yu, W, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-01-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
4OIM
 
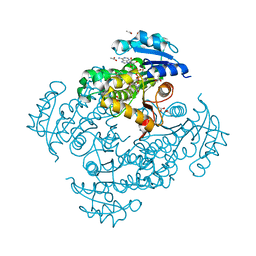 | | Crystal structure of Mycobacterium tuberculosis InhA in complex with inhibitor PT119 in 2.4 M acetate | | Descriptor: | 2-(2-CYANOPHENOXY)-5-HEXYLPHENOL, ACETATE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-01-20 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Time-Dependent Diaryl Ether Inhibitors of InhA: Structure-Activity Relationship Studies of Enzyme Inhibition, Antibacterial Activity, and in vivo Efficacy.
Chemmedchem, 9, 2014
|
|
6K6R
 
 | |
4OXK
 
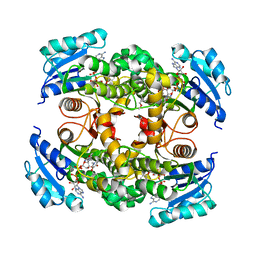 | | Multiple binding modes of inhibitor PT155 to the Mycobacterium tuberculosis enoyl-ACP reductase InhA within a tetramer | | Descriptor: | 3,6,9,12,15-pentaoxaoctadecan-17-amine, 5-(4-amino-2-methylphenoxy)-2-hexyl-4-hydroxy-1-methylpyridinium, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-02-05 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8429 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
4OYR
 
 | | Competition of the small inhibitor PT91 with large fatty acyl substrate of the Mycobacterium tuberculosis enoyl-ACP reductase InhA by induced substrate-binding loop refolding | | Descriptor: | 2-(2-chloranylphenoxy)-5-hexyl-phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-02-13 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2995 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
4OXY
 
 | | Substrate-binding loop movement with inhibitor PT10 in the tetrameric Mycobacterium tuberculosis enoyl-ACP reductase InhA | | Descriptor: | 5-hexyl-2-(2-nitrophenoxy)phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, H.J, Sullivan, T.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-02-09 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3501 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
4OXN
 
 | | Substrate-like binding mode of inhibitor PT155 to the Mycobacterium tuberculosis enoyl-ACP reductase InhA | | Descriptor: | 3,6,9,12,15-pentaoxaoctadecan-17-amine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-(4-amino-2-methylphenoxy)-2-hexyl-4-hydroxy-1-methylpyridinium, ... | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-02-05 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2926 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
6LR3
 
 | | Structural and functional insights into macrophage migration inhibitory factor from Oncomelania hupensis, the intermediate host of Schistosoma japonicum | | Descriptor: | Macrophage migration inhibitory factor, SULFATE ION | | Authors: | Su, Z.M, Tian, X.Y, Li, H.J, Wei, Z.M, Chen, L.F, Ren, H.X, Peng, W.F, Tang, C.T. | | Deposit date: | 2020-01-15 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and functional insights into macrophage migration inhibitory factor from Oncomelania hupensis, the intermediate host of Schistosoma japonicum.
Biochem.J., 477, 2020
|
|
5GJ9
 
 | | Crystal structure of Arabidopsis thaliana ACO2 in complex with POA | | Descriptor: | 1-aminocyclopropane-1-carboxylate oxidase 2, PYRAZINE-2-CARBOXYLIC ACID, ZINC ION | | Authors: | Sun, X.Z, Li, Y.X, He, W.R, Ji, C.G, Xia, P.X, Wang, Y.C, Du, S, Li, H.J, Raikhel, N, Xiao, J.Y, Guo, H.W. | | Deposit date: | 2016-06-28 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyrazinamide and derivatives block ethylene biosynthesis by inhibiting ACC oxidase.
Nat Commun, 8, 2017
|
|
5GJA
 
 | | Crystal structure of Arabidopsis thaliana ACO2 in complex with 2-PA | | Descriptor: | 1-aminocyclopropane-1-carboxylate oxidase 2, PYRIDINE-2-CARBOXYLIC ACID, ZINC ION | | Authors: | Sun, X.Z, Li, Y.X, He, W.R, Ji, C.G, Xia, P.X, Wang, Y.C, Du, S, Li, H.J, Raikhel, N, Xiao, J.Y, Guo, H.W. | | Deposit date: | 2016-06-28 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyrazinamide and derivatives block ethylene biosynthesis by inhibiting ACC oxidase.
Nat Commun, 8, 2017
|
|
6LKV
 
 | | Structural and functional insights into macrophage migration inhibitory factor from Oncomelania hupensis, the intermediate host of Schistosoma japonicum | | Descriptor: | CHLORIDE ION, Macrophage migration inhibitory factor, SULFATE ION | | Authors: | Su, Z.M, Tian, X.Y, Li, H.J, Wei, Z.M, Chen, L.F, Ren, H.X, Peng, W.F, Tang, C.T. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional insights into macrophage migration inhibitory factor from Oncomelania hupensis, the intermediate host of Schistosoma japonicum.
Biochem.J., 477, 2020
|
|
6LKW
 
 | | Structural and functional insights into macrophage migration inhibitory factor from Oncomelania hupensis, the intermediate host of Schistosoma japonicum | | Descriptor: | CHLORIDE ION, Macrophage migration inhibitory factor | | Authors: | Su, Z.M, Tian, X.Y, Li, H.J, Wei, Z.M, Chen, L.F, Ren, H.X, Peng, W.F, Tang, C.T. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and functional insights into macrophage migration inhibitory factor from Oncomelania hupensis, the intermediate host of Schistosoma japonicum.
Biochem.J., 477, 2020
|
|
