5MSS
 
 | |
5MSU
 
 | |
5MSD
 
 | |
5MSO
 
 | |
5MSW
 
 | |
5MSC
 
 | |
5MSP
 
 | |
5MSQ
 
 | |
5O3M
 
 | |
8AA7
 
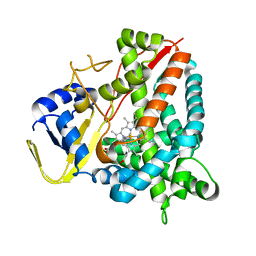 | | Crystal structure of a staphylococcal orthologue of CYP134A1 (CYPX) in complex with a heme coordinated fragment | | Descriptor: | 2,3,4,9-tetrahydro-1~{H}-pyrido[3,4-b]indole, Cytochrome P450 protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Snee, M, Katariya, M, Levy, C, Leys, D. | | Deposit date: | 2022-06-30 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a staphylococcal orthologue of CYP134A1 (CYPX) in complex with a heme coordinated fragment
To Be Published
|
|
2R14
 
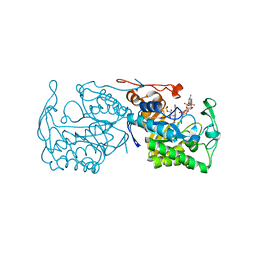 | | Structure of morphinone reductase in complex with tetrahydroNAD | | Descriptor: | 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN MONONUCLEOTIDE, Morphinone reductase | | Authors: | Costello, C.L, Scrutton, N.S, Leys, D. | | Deposit date: | 2007-08-22 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mutagenesis of morphinone reductase induces multiple reactive configurations and identifies potential ambiguity in kinetic analysis of enzyme tunneling mechanisms.
J.Am.Chem.Soc., 129, 2007
|
|
1SMJ
 
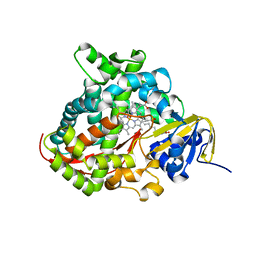 | | Structure of the A264E mutant of cytochrome P450 BM3 complexed with palmitoleate | | Descriptor: | Bifunctional P-450:NADPH-P450 reductase, PALMITOLEIC ACID, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Joyce, M.G, Girvan, H.M, Munro, A.W, Leys, D. | | Deposit date: | 2004-03-09 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A Single Mutation in Cytochrome P450 BM3 Induces the Conformational Rearrangement Seen upon Substrate Binding in the Wild-type Enzyme
J.Biol.Chem., 279, 2004
|
|
1T9G
 
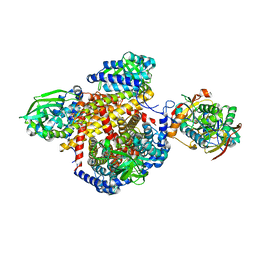 | | Structure of the human MCAD:ETF complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Acyl-CoA dehydrogenase, medium-chain specific, ... | | Authors: | Toogood, H.S, van Thiel, A, Basran, J, Sutcliffe, M.J, Scrutton, N.S, Leys, D. | | Deposit date: | 2004-05-17 | | Release date: | 2004-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Extensive domain motion and electron transfer in the human electron transferring flavoprotein-medium chain Acyl-CoA dehydrogenase complex
J.Biol.Chem., 279, 2004
|
|
2O2K
 
 | | Crystal Structure of the Activation Domain of Human Methionine Synthase Isoform/Mutant D963E/K1071N | | Descriptor: | Methionine synthase | | Authors: | Wolthers, K.R, Toogood, H.S, Jowitt, T.A, Marshall, K.R, Leys, D, Scrutton, N.S. | | Deposit date: | 2006-11-30 | | Release date: | 2006-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure and solution characterization of the activation domain of human methionine synthase
Febs J., 274, 2007
|
|
3CLR
 
 | | Crystal structure of the R236A ETF mutant from M. methylotrophus | | Descriptor: | ADENOSINE MONOPHOSPHATE, Electron transfer flavoprotein subunit alpha, Electron transfer flavoprotein subunit beta, ... | | Authors: | Katona, G, Leys, D. | | Deposit date: | 2008-03-20 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the dynamic interface between trimethylamine dehydrogenase (TMADH) and electron transferring flavoprotein (ETF) in the TMADH-2ETF complex: role of the Arg-alpha237 (ETF) and Tyr-442 (TMADH) residue pair.
Biochemistry, 47, 2008
|
|
5N6G
 
 | | NerA from Agrobacterium radiobacter in complex with 2-phenylacrylic acid | | Descriptor: | 2-Phenylacrylic acid, ACETATE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Karuppiah, V, Toogood, H.S, Leys, D, Scrutton, N.S. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural insights into the ene-reductase synthesis of profens.
Org. Biomol. Chem., 15, 2017
|
|
5N6Q
 
 | | Xenobiotic reductase A (XenA) from Pseudomonas putida in complex with 2-phenylacrylic acid | | Descriptor: | 2-Phenylacrylic acid, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Karuppiah, V, Toogood, H.S, Leys, D, Scrutton, N.S. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the ene-reductase synthesis of profens.
Org. Biomol. Chem., 15, 2017
|
|
8J2W
 
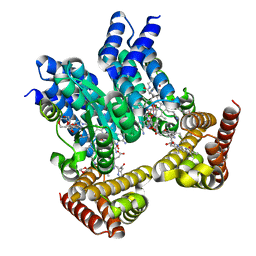 | | Saccharothrix syringae photocobilins protein, dark state | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 5'-DEOXYADENOSINE, BILIVERDINE IX ALPHA, ... | | Authors: | Zhang, S, Poddar, H, Levy, W.C, Leys, D. | | Deposit date: | 2023-04-15 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Photocobilins integrate B12 and bilin photochemistry for enzyme control.
Nat Commun, 15, 2024
|
|
8J2Y
 
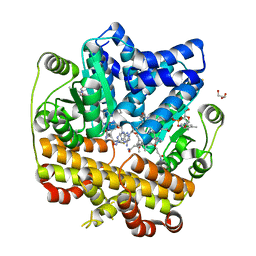 | | Acidimicrobiaceae bacterium photocobilins protein, dark state | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, S, Poddar, H, Levy, W.C, Leys, D. | | Deposit date: | 2023-04-15 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Photocobilins integrate B12 and bilin photochemistry for enzyme control.
Nat Commun, 15, 2024
|
|
8J2X
 
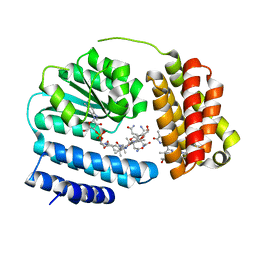 | | Saccharothrix syringae photocobilins protein, light state | | Descriptor: | BILIVERDINE IX ALPHA, COBALAMIN, Cobalamin-binding protein, ... | | Authors: | Zhang, S, Poddar, H, Levy, C, Leys, D. | | Deposit date: | 2023-04-15 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Photocobilins integrate B12 and bilin photochemistry for enzyme control.
Nat Commun, 15, 2024
|
|
5O3N
 
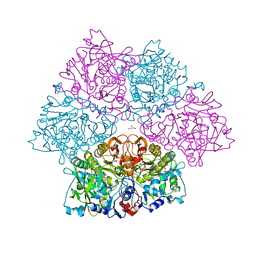 | | Crystal structure of E. cloacae 3,4-dihydroxybenzoic acid decarboxylase (AroY) reconstituted with prFMN | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, 3,4-dihydroxybenzoate decarboxylase, GLYCEROL, ... | | Authors: | Marshall, S.A, Leys, D. | | Deposit date: | 2017-05-24 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Regioselective para-Carboxylation of Catechols with a Prenylated Flavin Dependent Decarboxylase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
2QTZ
 
 | | Crystal Structure of the NADP+-bound FAD-containing FNR-like Module of Human Methionine Synthase Reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Methionine synthase reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wolthers, K.R, Lou, X, Toogood, H.S, Leys, D, Scrutton, N.S. | | Deposit date: | 2007-08-02 | | Release date: | 2007-11-13 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of Coenzyme Binding to Human Methionine Synthase Reductase Revealed through the Crystal Structure of the FNR-like Module and Isothermal Titration Calorimetry
Biochemistry, 46, 2007
|
|
3CLU
 
 | | Crystal structure of the R236K mutant from Methylophilus methylotrophus ETF | | Descriptor: | ADENOSINE MONOPHOSPHATE, Electron transfer flavoprotein subunit alpha, Electron transfer flavoprotein subunit beta, ... | | Authors: | Katona, G, Leys, D. | | Deposit date: | 2008-03-20 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the dynamic interface between trimethylamine dehydrogenase (TMADH) and electron transferring flavoprotein (ETF) in the TMADH-2ETF complex: role of the Arg-alpha237 (ETF) and Tyr-442 (TMADH) residue pair.
Biochemistry, 47, 2008
|
|
1JRX
 
 | | Crystal structure of Arg402Ala mutant flavocytochrome c3 from Shewanella frigidimarina | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FLAVOCYTOCHROME C, FUMARIC ACID, ... | | Authors: | Mowat, C.G, Moysey, R, Miles, C.S, Leys, D, Doherty, M.K, Taylor, P, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2001-08-15 | | Release date: | 2001-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and crystallographic analysis of the key active site acid/base arginine in a soluble fumarate reductase.
Biochemistry, 40, 2001
|
|
3CLT
 
 | | Crystal structure of the R236E mutant of Methylophilus methylotrophus ETF | | Descriptor: | ADENOSINE MONOPHOSPHATE, Electron transfer flavoprotein subunit alpha, Electron transfer flavoprotein subunit beta, ... | | Authors: | Katona, G, Leys, D. | | Deposit date: | 2008-03-20 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the dynamic interface between trimethylamine dehydrogenase (TMADH) and electron transferring flavoprotein (ETF) in the TMADH-2ETF complex: role of the Arg-alpha237 (ETF) and Tyr-442 (TMADH) residue pair.
Biochemistry, 47, 2008
|
|
