8C37
 
 | |
8C32
 
 | |
7AE5
 
 | |
8C31
 
 | |
8C34
 
 | |
7NF3
 
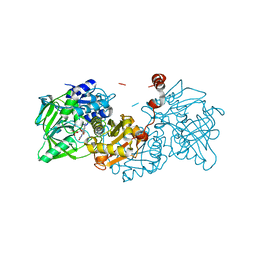 | | Structure of A. niger Fdc T395M variant (AnFdcI) in complex with prFMN | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Ferulic acid decarboxylase 1, MANGANESE (II) ION, ... | | Authors: | Saaret, A, Leys, D. | | Deposit date: | 2021-02-05 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Directed evolution of prenylated FMN-dependent Fdc supports efficient in vivo isobutene production.
Nat Commun, 12, 2021
|
|
7NEY
 
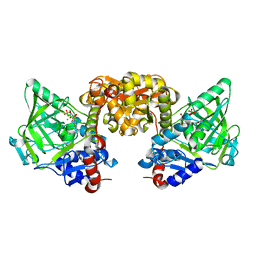 | | Structure of T. atroviride Fdc wild-type (TaFdc) in complex with prFMN | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Ferulic acid decarboxylase 1, MANGANESE (II) ION, ... | | Authors: | Saaret, A, Leys, D. | | Deposit date: | 2021-02-05 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Directed evolution of prenylated FMN-dependent Fdc supports efficient in vivo isobutene production.
Nat Commun, 12, 2021
|
|
3CLR
 
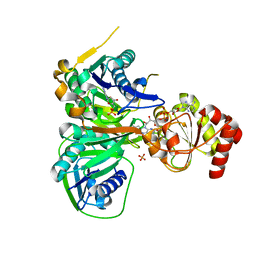 | | Crystal structure of the R236A ETF mutant from M. methylotrophus | | Descriptor: | ADENOSINE MONOPHOSPHATE, Electron transfer flavoprotein subunit alpha, Electron transfer flavoprotein subunit beta, ... | | Authors: | Katona, G, Leys, D. | | Deposit date: | 2008-03-20 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the dynamic interface between trimethylamine dehydrogenase (TMADH) and electron transferring flavoprotein (ETF) in the TMADH-2ETF complex: role of the Arg-alpha237 (ETF) and Tyr-442 (TMADH) residue pair.
Biochemistry, 47, 2008
|
|
7ZQR
 
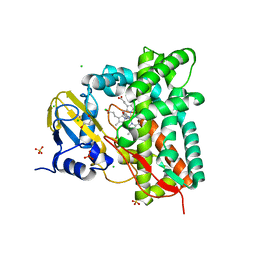 | | Crystal structure of CYP125 from Mycobacterium tuberculosis in complex with an inhibitor | | Descriptor: | 4-(4-methoxyphenyl)pyridine, CHLORIDE ION, GLYCEROL, ... | | Authors: | Snee, M, Katariya, M, Tunnicliffe, R, Levy, C, Leys, D. | | Deposit date: | 2022-05-02 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis.
Chemistry, 29, 2023
|
|
7ZSU
 
 | | Crystal structure of CYP125 from Mycobacterium tuberculosis in complex with an inhibitor | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, Steroid C26-monooxygenase, ... | | Authors: | Snee, M, Katariya, M, Levy, C, Leys, D. | | Deposit date: | 2022-05-09 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis.
Chemistry, 29, 2023
|
|
7ZXD
 
 | | Crystal structure of CYP125 from Mycobacterium tuberculosis in complex with an inhibitor | | Descriptor: | 1-[1-(2-piperidin-4-ylethyl)-5-pyridin-4-yl-indol-2-yl]butan-1-one, CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Snee, M, Katariya, M, Levy, C, Leys, D. | | Deposit date: | 2022-05-20 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis.
Chemistry, 29, 2023
|
|
7ZT0
 
 | | Crystal structure of CYP125 from Mycobacterium tuberculosis in complex with an inhibitor | | Descriptor: | 1-(2-piperazin-1-ylethyl)-5-pyridin-4-yl-indole-2-carboxamide, CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Snee, M, Katariya, M, Levy, C, Leys, D. | | Deposit date: | 2022-05-09 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis.
Chemistry, 29, 2023
|
|
7AE4
 
 | |
2Q7Q
 
 | |
8CRD
 
 | |
7AE7
 
 | |
2R14
 
 | | Structure of morphinone reductase in complex with tetrahydroNAD | | Descriptor: | 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN MONONUCLEOTIDE, Morphinone reductase | | Authors: | Costello, C.L, Scrutton, N.S, Leys, D. | | Deposit date: | 2007-08-22 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mutagenesis of morphinone reductase induces multiple reactive configurations and identifies potential ambiguity in kinetic analysis of enzyme tunneling mechanisms.
J.Am.Chem.Soc., 129, 2007
|
|
6FP0
 
 | |
6FOX
 
 | |
2QTZ
 
 | | Crystal Structure of the NADP+-bound FAD-containing FNR-like Module of Human Methionine Synthase Reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Methionine synthase reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wolthers, K.R, Lou, X, Toogood, H.S, Leys, D, Scrutton, N.S. | | Deposit date: | 2007-08-02 | | Release date: | 2007-11-13 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of Coenzyme Binding to Human Methionine Synthase Reductase Revealed through the Crystal Structure of the FNR-like Module and Isothermal Titration Calorimetry
Biochemistry, 46, 2007
|
|
7NF4
 
 | |
7NF0
 
 | |
7NF1
 
 | |
7NF2
 
 | |
6H6V
 
 | |
