5MK3
 
 | |
5MK0
 
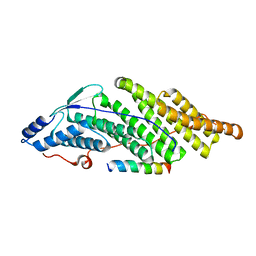 | |
5LM1
 
 | | Crystal Structure of HD-PTP phosphatase in complex with UBAP1 | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 23, UBAP-1 | | Authors: | Levy, C. | | Deposit date: | 2016-07-28 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis for Selective Interaction between the ESCRT Regulator HD-PTP and UBAP1.
Structure, 24, 2016
|
|
5MK1
 
 | |
5MK2
 
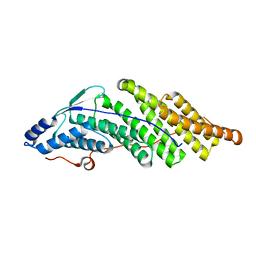 | |
5LM2
 
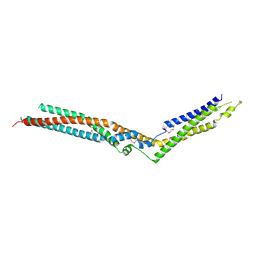 | | Crystal Structure of HD-PTP phosphatase | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 23 | | Authors: | Levy, C. | | Deposit date: | 2016-07-28 | | Release date: | 2016-11-30 | | Last modified: | 2016-12-14 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural Basis for Selective Interaction between the ESCRT Regulator HD-PTP and UBAP1.
Structure, 24, 2016
|
|
3IA4
 
 | |
6Y2V
 
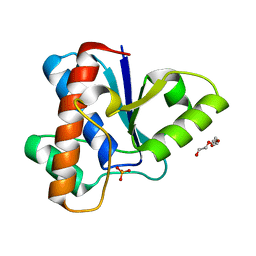 | |
6Y2W
 
 | |
7NDS
 
 | | Crystal structure of TphC in a closed conformation | | Descriptor: | Tripartite tricarboxylate transporter substrate binding protein, terephthalic acid | | Authors: | Levy, C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of terephthalate recognition by solute binding protein TphC.
Nat Commun, 12, 2021
|
|
7NDR
 
 | | Crystal structure of TphC in an open conformation | | Descriptor: | 1,2-ETHANEDIOL, Tripartite tricarboxylate transporter substrate binding protein | | Authors: | Levy, C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis of terephthalate recognition by solute binding protein TphC.
Nat Commun, 12, 2021
|
|
7YXF
 
 | | Crystal structure of CYP125 from Mycobacterium tuberculosis in complex with an inhibitor | | Descriptor: | 1-(2-piperazin-1-ylethyl)-5-pyridin-4-yl-indole, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Snee, M, Katariya, M, Levy, C, Leys, D. | | Deposit date: | 2022-02-16 | | Release date: | 2023-03-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis.
Chemistry, 29, 2023
|
|
7ZP7
 
 | | Crystal structure of evolved photoenzyme EnT1.3 (truncated) with bound product | | Descriptor: | (1~{R},10~{R},12~{S})-15-oxa-8-azatetracyclo[8.5.0.0^{1,12}.0^{2,7}]pentadeca-2(7),3,5-trien-9-one, 1,2-ETHANEDIOL, EnT1.3 C | | Authors: | Hardy, F.J, Levy, C. | | Deposit date: | 2022-04-26 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A designed photoenzyme for enantioselective [2+2] cycloadditions.
Nature, 611, 2022
|
|
8J2X
 
 | | Saccharothrix syringae photocobilins protein, light state | | Descriptor: | BILIVERDINE IX ALPHA, COBALAMIN, Cobalamin-binding protein, ... | | Authors: | Zhang, S, Poddar, H, Levy, C, Leys, D. | | Deposit date: | 2023-04-15 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Photocobilins integrate B12 and bilin photochemistry for enzyme control.
Nat Commun, 15, 2024
|
|
5EDT
 
 | | Crystal structure of Mycobacterium tuberculosis CYP121 in complex with LIG9 | | Descriptor: | 2-azanyl-5-chloranyl-benzamide, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kavanagh, M.E, Coyne, A.G, McLean, K.J, James, G.G, Levy, C, Marino, L.B, Carvalho, L.P.D, Chan, D.S.H, Hudson, S.A, Surade, S, Munro, A.W, Abell, C. | | Deposit date: | 2015-10-22 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Fragment-Based Approaches to the Development of Mycobacterium tuberculosis CYP121 Inhibitors.
J.Med.Chem., 59, 2016
|
|
8C3W
 
 | | Crystal structure of a computationally designed heme binding protein, dnHEM1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ortmayer, M, Levy, C. | | Deposit date: | 2022-12-29 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of Heme Enzymes with a Tunable Substrate Binding Pocket Adjacent to an Open Metal Coordination Site.
J.Am.Chem.Soc., 145, 2023
|
|
6RQ5
 
 | | CYP121 in complex with 3,5-dimethyl dicyclotyrosine | | Descriptor: | (3~{S},6~{S})-3-[(3,5-dimethyl-4-oxidanyl-phenyl)methyl]-6-[(4-hydroxyphenyl)methyl]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mycocyclosin synthase, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Activity Relationships of cyclo (l-Tyrosyl-l-tyrosine) Derivatives Binding to Mycobacterium tuberculosis CYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
6RQ3
 
 | | CYP121 in complex with 2,6-dimethyl dicyclotyrosine | | Descriptor: | (3~{S},6~{S})-3-[(2,6-dimethyl-4-oxidanyl-phenyl)methyl]-6-[(4-hydroxyphenyl)methyl]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mycocyclosin synthase, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Activity Relationships of cyclo (l-Tyrosyl-l-tyrosine) Derivatives Binding to Mycobacterium tuberculosis CYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
6RQE
 
 | | CYP121 in complex with 3-acetylene dicyclotyrosine | | Descriptor: | 3-acetylene dicyclotyrosine, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structure-Activity Relationships ofcyclo(l-Tyrosyl-l-tyrosine) Derivatives Binding toMycobacterium tuberculosisCYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
6RQ8
 
 | | CYP121 in complex with 3-iodo dicyclotyrosine | | Descriptor: | (3~{S},6~{S})-3-[(4-hydroxyphenyl)methyl]-6-[(3-iodanyl-4-oxidanyl-phenyl)methyl]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mycocyclosin synthase, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structure-Activity Relationships of cyclo (l-Tyrosyl-l-tyrosine) Derivatives Binding to Mycobacterium tuberculosis CYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
2CG8
 
 | | The bifunctional dihydroneopterin aldolase 6-hydroxymethyl-7,8- dihydropterin synthase from Streptococcus pneumoniae | | Descriptor: | DIHYDRONEOPTERIN ALDOLASE 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN SYNTHASE | | Authors: | Garcon, A, Levy, C, Derrick, J.P. | | Deposit date: | 2006-02-28 | | Release date: | 2006-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Bifunctional Dihydroneopterin Aldolase/6-Hydroxymethyl-7,8-Dihydropterin Pyrophosphokinase from Streptococcus Pneumoniae.
J.Mol.Biol., 360, 2006
|
|
2VEG
 
 | |
2VEF
 
 | | Dihydropteroate synthase from Streptococcus pneumoniae | | Descriptor: | DIHYDROPTEROATE SYNTHASE, PHOSPHATE ION | | Authors: | Derrick, J.P, Levy, C. | | Deposit date: | 2007-10-24 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dihydropteroate Synthase from Streptococcus Pneumoniae: Structure, Ligand Recognition and Mechanism of Sulfonamide Resistance.
Biochem.J., 412, 2008
|
|
6HAU
 
 | | KSHV PAN RNA Mta-response element fragment complexed with the globular domain of herpesvirus saimiri ORF57 | | Descriptor: | ACETATE ION, MRE fragment of PAN RNA, ZINC ION, ... | | Authors: | Tunnicliffe, R.B, Levy, C, Ruiz Nivia, H.D, Sandri-Goldin, R.M, Golovanov, A.P. | | Deposit date: | 2018-08-08 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural identification of conserved RNA binding sites in herpesvirus ORF57 homologs: implications for PAN RNA recognition.
Nucleic Acids Res., 47, 2019
|
|
6HAT
 
 | | Globular domain of herpesvirus saimiri ORF57 | | Descriptor: | ACETATE ION, ZINC ION, mRNA export factor ICP27 homolog | | Authors: | Tunnicliffe, R.B, Levy, C, Ruiz Nivia, H.D, Sandri-Goldin, R.M, Golovanov, A.P. | | Deposit date: | 2018-08-08 | | Release date: | 2018-11-21 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (1.856 Å) | | Cite: | Structural identification of conserved RNA binding sites in herpesvirus ORF57 homologs: implications for PAN RNA recognition.
Nucleic Acids Res., 47, 2019
|
|
