5ZHJ
 
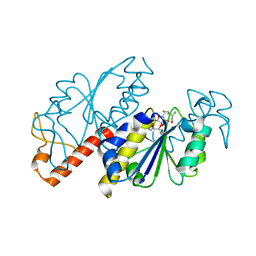 | | Crystal structure of TrmD from Mycobacterium tuberculosis in complex with S-adenosyl homocysteine (SAH) | | Descriptor: | GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Zhong, W, Pasunooti, K.K, Balamkundu, S, Wong, Y.W, Nah, Q, Liu, C.F, Lescar, J, Dedon, P.C. | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Thienopyrimidinone Derivatives That Inhibit Bacterial tRNA (Guanine37-N1)-Methyltransferase (TrmD) by Restructuring the Active Site with a Tyrosine-Flipping Mechanism.
J.Med.Chem., 62, 2019
|
|
5ZBI
 
 | |
2J7W
 
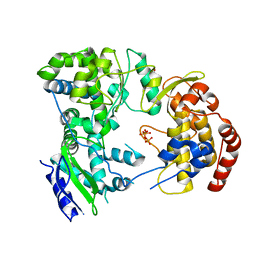 | | Dengue virus NS5 RNA dependent RNA polymerase domain complexed with 3' dGTP | | Descriptor: | DI(HYDROXYETHYL)ETHER, GUANOSINE-5'-TRIPHOSPHATE, POLYPROTEIN, ... | | Authors: | Yap, T.L, Xu, T, Chen, Y.L, Malet, H, Egloff, M.P, Canard, B, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2006-10-17 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Dengue Virus RNA- Dependent RNA Polymerase Catalytic Domain at 1.85 Angstrom Resolution.
J.Virol., 81, 2007
|
|
2J7U
 
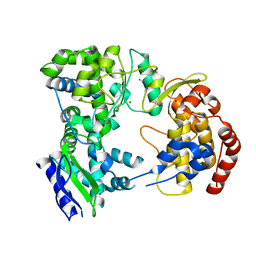 | | Dengue virus NS5 RNA dependent RNA polymerase domain | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, RNA DEPENDENT RNA POLYMERASE, ... | | Authors: | Yap, T.L, Xu, T, Chen, Y.L, Malet, H, Egloff, M.P, Canard, B, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2006-10-17 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Dengue Virus RNA- Dependent RNA Polymerase Catalytic Domain at 1.85 Angstrom Resolution.
J.Virol., 81, 2007
|
|
7DU3
 
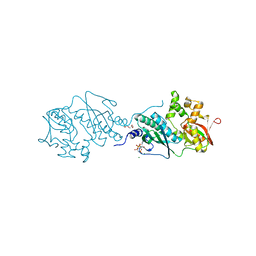 | | ThiL in complex with AMP-PNP | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Lin, J.Q, Lescar, J. | | Deposit date: | 2021-01-07 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | ThiL in complex with AMP-PNP
To Be Published
|
|
7F8C
 
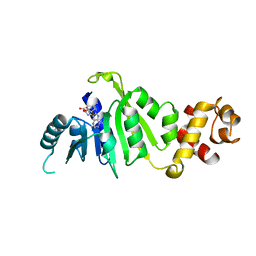 | |
7F8A
 
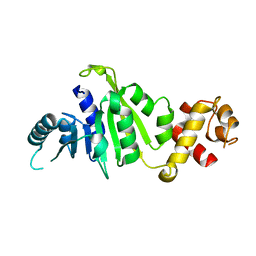 | | Crystal structure of rRNA methyltransferase Erm38 | | Descriptor: | Erm(38) | | Authors: | Goh, B.C, Lescar, J. | | Deposit date: | 2021-07-01 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and functional analysis of mycobacterial erythromycin resistance methyltransferase Erm38 reveals its RNA-binding site.
J.Biol.Chem., 298, 2022
|
|
7F8B
 
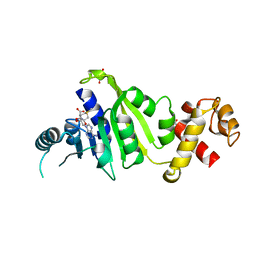 | |
7F5P
 
 | | The crystal structure of VyPAL2-C214A, a dead mutant of VyPAL2 peptide asparaginyl ligase in form I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Peptide Asparaginyl Ligases, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, S, Sahili, A, Lescar, J. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for proenzyme maturation, substrate recognition, and ligation by a hyperactive peptide asparaginyl ligase.
Plant Cell, 34, 2022
|
|
7F5Q
 
 | | The crystal structure of VyPAL2 peptide asparaginyl ligase in its active enzyme form | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, S, Sahili, A, Lescar, J. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for proenzyme maturation, substrate recognition, and ligation by a hyperactive peptide asparaginyl ligase.
Plant Cell, 34, 2022
|
|
7F5J
 
 | | The crystal structure of VyPAL2-I244V, a more efficient mutant of VyPAL2 peptide asparaginyl ligase in its active enzyme form | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, S, Sahili, A, Lescar, J. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.593 Å) | | Cite: | Structural basis for proenzyme maturation, substrate recognition, and ligation by a hyperactive peptide asparaginyl ligase.
Plant Cell, 34, 2022
|
|
7FA0
 
 | | The crystal structure of VyPAL2-C214A, a dead mutant of VyPAL2 peptide asparaginyl ligase in form II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Peptide Asparaginyl Ligases, ... | | Authors: | Hu, S, Sahili, A, Lescar, J. | | Deposit date: | 2021-07-05 | | Release date: | 2022-07-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for proenzyme maturation, substrate recognition, and ligation by a hyperactive peptide asparaginyl ligase.
Plant Cell, 34, 2022
|
|
