6L6I
 
 | | hASIC1a co-crystallized with Mamb-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1 | | Authors: | Lei, F, Jian, S. | | Deposit date: | 2019-10-29 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | hASIC1a co-crystallized with Mamb-1
To Be Published
|
|
6L6N
 
 | | hASIC1a co-crystallized with Nafamostat | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lei, F, Jian, S. | | Deposit date: | 2019-10-29 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | hASIC1a co-crystallized with Mamb-1
To Be Published
|
|
6L6P
 
 | |
7Q8E
 
 | |
7NED
 
 | |
6XTS
 
 | |
6XTR
 
 | |
6XTQ
 
 | |
6XTO
 
 | |
6XTP
 
 | |
6XTN
 
 | |
6XTL
 
 | |
6S7Q
 
 | |
6S7J
 
 | |
4XGC
 
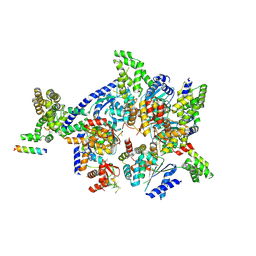 | | Crystal structure of the eukaryotic origin recognition complex | | Descriptor: | CHLORIDE ION, Origin recognition complex subunit 1, Origin recognition complex subunit 2, ... | | Authors: | Bleichert, F, Botchan, M.R, Berger, J.M. | | Deposit date: | 2014-12-30 | | Release date: | 2015-04-01 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the eukaryotic origin recognition complex.
Nature, 519, 2015
|
|
6XTM
 
 | |
6S07
 
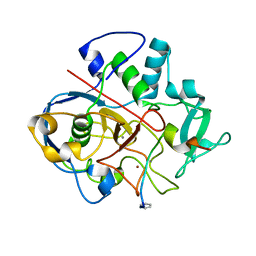 | | Structure of formylglycine-generating enzyme at 1.04 A in complex with copper and substrate reveals an acidic pocket for binding and acti-vation of molecular oxygen. | | Descriptor: | Abz-ALA-THR-THR-PRO-LEU-CYS-GLY-PRO-SER-ARG-ALA-SER-ILE-LEU-SER-GLY-ARG, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Leisinger, F, Miarzlou, D.A, Seebeck, F.P. | | Deposit date: | 2019-06-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Structure of formylglycine-generating enzyme in complex with copper and a substrate reveals an acidic pocket for binding and activation of molecular oxygen.
Chem Sci, 10, 2019
|
|
8SIU
 
 | |
7ZAY
 
 | | Human heparan sulfate polymerase complex EXT1-EXT2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Exostosin-1, Exostosin-2, ... | | Authors: | Leisico, F, Omeiri, J, Hons, M, Schoehn, G, Lortat-Jacob, H, Wild, R. | | Deposit date: | 2022-03-23 | | Release date: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the human heparan sulfate polymerase complex EXT1-EXT2.
Nat Commun, 13, 2022
|
|
8SIY
 
 | |
3MX8
 
 | | Crystal structure of ribonuclease A tandem enzymes and their interaction with the cytosolic ribonuclease inhibitor | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, LINKER, ... | | Authors: | Leich, F, Neumann, P, Lilie, H, Ulbrich-Hofmann, R, Arnold, U. | | Deposit date: | 2010-05-07 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of RNase A tandem enzymes and their interaction with the cytosolic ribonuclease inhibitor
Febs J., 278, 2011
|
|
8AJP
 
 | |
4ISV
 
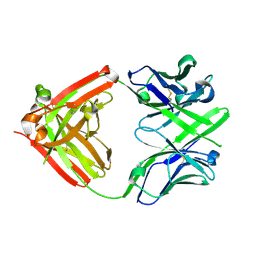 | | Crystal structure of the Fab FRAGMENT OF 1C2, A MONOCLONAL ANTIBODY SPECIFIC FOR POLY-GLUTAMINE | | Descriptor: | 1C2 FAB HEAVY CHAIN, 1C2 FAB LIGHT CHAIN | | Authors: | Klein, F.A.C, Zeder-Lutz, G, Cousido-Siah, A, Mitschler, A, Katz, A, Eberling, P, Mandel, J.L, Podjarny, A, Trottier, Y. | | Deposit date: | 2013-01-17 | | Release date: | 2013-07-03 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Linear and extended: a common polyglutamine conformation recognized by the three antibodies MW1, 1C2 and 3B5H10.
Hum.Mol.Genet., 22, 2013
|
|
4JJ5
 
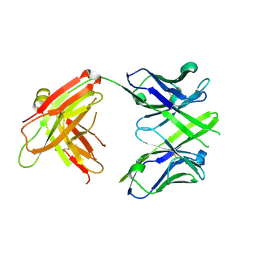 | | CRYSTAL STRUCTURE OF THE Fab FRAGMENT OF 1C2, A MONOCLONAL ANTIBODY SPECIFIC for POLY-GLUTAMINE | | Descriptor: | 1C2 FAB HEAVY CHAIN, 1C2 FAB LIGHT CHAIN | | Authors: | Klein, F.A.C, Zeder-Lutz, G, Cousido-Siah, A, Mitschler, A, Katz, A, Eberling, P, Mandel, J.L, Podjarny, A, Trottier, Y. | | Deposit date: | 2013-03-07 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Linear and extended: a common polyglutamine conformation recognized by the three antibodies MW1, 1C2 and 3B5H10.
Hum.Mol.Genet., 22, 2013
|
|
6YIR
 
 | |
