1YN1
 
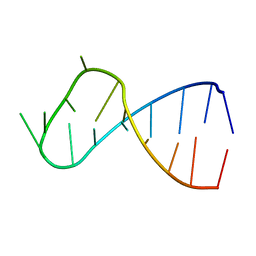 | |
3IVN
 
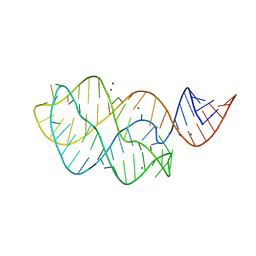 | | Structure of the U65C mutant A-riboswitch aptamer from the Bacillus subtilis pbuE operon | | Descriptor: | A-riboswitch, BROMIDE ION, MAGNESIUM ION | | Authors: | Delfosse, V, Dagenais, P, Chausse, D, Di Tomasso, G, Legault, P. | | Deposit date: | 2009-09-01 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Riboswitch structure: an internal residue mimicking the purine ligand.
Nucleic Acids Res., 38, 2010
|
|
2GS0
 
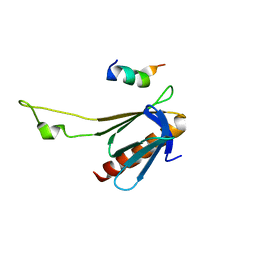 | | NMR structure of the complex between the PH domain of the Tfb1 subunit from TFIIH and the activation domain of p53 | | Descriptor: | Cellular tumor antigen p53, RNA polymerase II transcription factor B subunit 1 | | Authors: | Di Lello, P, Jones, T.N, Nguyen, B.D, Legault, P, Omichinski, J.G. | | Deposit date: | 2006-04-25 | | Release date: | 2006-10-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the Tfb1/p53 complex: Insights into the interaction between the p62/Tfb1 subunit of TFIIH and the activation domain of p53.
Mol.Cell, 22, 2006
|
|
1S6L
 
 | | Solution structure of MerB, the Organomercurial Lyase involved in the bacterial mercury resistance system | | Descriptor: | Alkylmercury lyase | | Authors: | Di Lello, P, Benison, G.C, Valafar, H, Pitts, K.E, Summers, A.O, Legault, P, Omichinski, J.G. | | Deposit date: | 2004-01-25 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structural studies reveal a novel protein fold for MerB, the organomercurial lyase involved in the bacterial mercury resistance system.
Biochemistry, 43, 2004
|
|
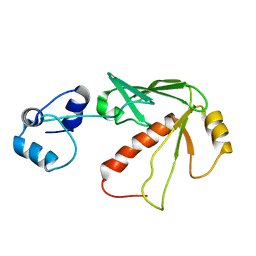
1YN2
 
 | |
5URN
 
 | | NMR structure of the complex between the PH domain of the Tfb1 subunit from TFIIH and the transactivation domain 1 of p65 | | Descriptor: | RNA polymerase II transcription factor B subunit 1, Transcription factor p65 | | Authors: | Lecoq, L, Omichinski, J.G, Raiola, L, Cyr, N, Chabot, P, Arseneault, G, Legault, P. | | Deposit date: | 2017-02-11 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of interactions between transactivation domain 1 of the p65 subunit of NF-kappa B and transcription regulatory factors.
Nucleic Acids Res., 45, 2017
|
|
1BNB
 
 | |
1DL6
 
 | | SOLUTION STRUCTURE OF HUMAN TFIIB N-TERMINAL DOMAIN | | Descriptor: | TRANSCRIPTION FACTOR II B (TFIIB), ZINC ION | | Authors: | Chen, H.-T, Legault, P, Glushka, J, Omichinski, J.G, Scott, R.A. | | Deposit date: | 1999-12-08 | | Release date: | 2000-10-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a (Cys3His) zinc ribbon, a ubiquitous motif in archaeal and eucaryal transcription.
Protein Sci., 9, 2000
|
|
1Y5O
 
 | | NMR structure of the amino-terminal domain from the Tfb1 subunit of yeast TFIIH | | Descriptor: | RNA polymerase II transcription factor B 73 kDa subunit | | Authors: | Di Lello, P, Nguyen, B.D, Jones, T.N, Potempa, K, Kobor, M.S, Legault, P, Omichinski, J.G. | | Deposit date: | 2004-12-02 | | Release date: | 2005-05-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Amino-Terminal Domain from the Tfb1 Subunit of TFIIH and
Characterization of Its Phosphoinositide and VP16 Binding Sites
Biochemistry, 44, 2005
|
|
1LDZ
 
 | |
1OW9
 
 | | NMR Structure of the Active Conformation of the VS Ribozyme Cleavage Site | | Descriptor: | A mimic of the VS Ribozyme Hairpin Substrate | | Authors: | Hoffmann, B, Mitchell, G.T, Gendron, P, Major, F, Andersen, A.A, Collins, R.A, Legault, P. | | Deposit date: | 2003-03-28 | | Release date: | 2003-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Active Conformation of the Varkud satellite Ribozyme Cleavage Site
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1TBK
 
 | |
1NHA
 
 | | Solution Structure of the Carboxyl-Terminal Domain of RAP74 and NMR Characterization of the FCP-Binding Sites of RAP74 and CTD of RAP74, the subunit of Human TFIIF | | Descriptor: | Transcription initiation factor IIF, alpha subunit | | Authors: | Nguyen, B.D, Chen, H.T, Kobor, M.S, Greenblatt, J, Legault, P, Omichinski, J.G. | | Deposit date: | 2002-12-19 | | Release date: | 2003-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Carboxyl-Terminal Domain of RAP74 and NMR Characterization of the FCP1-Binding Sites of RAP74 and Human TFIIB.
Biochemistry, 42, 2003
|
|
1ONV
 
 | | NMR Structure of a Complex Containing the TFIIF Subunit RAP74 and the RNAP II CTD Phosphatase FCP1 | | Descriptor: | Transcription initiation factor IIF, alpha subunit, serine phosphatase FCP1a | | Authors: | Nguyen, B.D, Abbott, K.L, Potempa, K, Kobor, M.S, Archambault, J, Greenblatt, J, Legault, P, Omichinski, J.G. | | Deposit date: | 2003-03-02 | | Release date: | 2003-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of a Complex Containing the TFIIF Subunit RAP74 and the RNA polymerase II carboxyl-terminal domain phosphatase FCP1
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2LDZ
 
 | |
2L5Z
 
 | | NMR structure of the A730 loop of the Neurospora VS ribozyme | | Descriptor: | RNA (26-MER) | | Authors: | Desjardins, G, Bonneau, E, Girard, N, Boisbouvier, J, Legault, P. | | Deposit date: | 2010-11-10 | | Release date: | 2011-02-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the A730 loop of the Neurospora VS ribozyme: insights into the formation of the active site.
Nucleic Acids Res., 39, 2011
|
|
2KDT
 
 | | PC1/3 DCSG sorting domain structure in DPC | | Descriptor: | Neuroendocrine convertase 1 | | Authors: | Dikeakos, J.D, Di Lello, P, Lacombe, M.J, Ghirlando, R, Legault, P, Reudelhuber, T.L, Omichinski, J.G. | | Deposit date: | 2009-01-19 | | Release date: | 2009-04-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Functional and structural characterization of a dense core secretory granule sorting domain from the PC1/3 protease.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2K7L
 
 | | NMR structure of a complex formed by the C-terminal domain of human RAP74 and a phosphorylated peptide from the central domain of the FCP1 | | Descriptor: | General transcription factor IIF subunit 1, centFCP1-T584PO4 peptide | | Authors: | Yang, A, Abbott, K.L, Desjardins, A, Di Lello, P, Omichinski, J.G, Legault, P. | | Deposit date: | 2008-08-13 | | Release date: | 2009-06-02 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a complex formed by the carboxyl-terminal domain of human RAP74 and a phosphorylated peptide from the central domain of the FCP1 phosphatase
Biochemistry, 48, 2009
|
|
2MI0
 
 | | NMR structure of the I-V kissing-loop interaction of the Neurospora VS ribozyme | | Descriptor: | 5'-R(*GP*AP*GP*CP*AP*GP*CP*AP*UP*CP*GP*UP*CP*GP*GP*CP*UP*GP*CP*UP*CP*A)-3', 5'-R(*GP*CP*GP*GP*CP*AP*GP*UP*UP*GP*AP*CP*UP*AP*CP*UP*GP*UP*CP*GP*C)-3' | | Authors: | Bouchard, P, Legault, P. | | Deposit date: | 2013-12-05 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insights into substrate recognition by the neurospora varkud satellite ribozyme: importance of u-turns at the kissing-loop junction.
Biochemistry, 53, 2014
|
|
2KE3
 
 | | PC1/3 DCSG sorting domain in CHAPS | | Descriptor: | Neuroendocrine convertase 1 | | Authors: | Dikeakos, J.D, Di Lello, P, Lacombe, M.J, Ghirlando, R, Legault, P, Reudelhuber, T.L, Omichinski, J.G. | | Deposit date: | 2009-01-22 | | Release date: | 2009-04-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Functional and structural characterization of a dense core secretory granule sorting domain from the PC1/3 protease
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2M14
 
 | | NMR structure of the complex between the PH domain of the Tfb1 subunit from TFIIH and Rad4 | | Descriptor: | DNA repair protein RAD4, RNA polymerase II transcription factor B subunit 1 | | Authors: | Lafrance-Vanasse, J, Arseneault, G, Cappadocia, L, Legault, P, Omichinski, J.G. | | Deposit date: | 2012-11-16 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and functional evidence that Rad4 competes with Rad2 for binding to the Tfb1 subunit of TFIIH in NER.
Nucleic Acids Res., 41, 2013
|
|
2MTK
 
 | |
2N3R
 
 | | NMR structure of the II-III-VI three-way junction from the VS ribozyme and identification of magnesium-binding sites using paramagnetic relaxation enhancement | | Descriptor: | MAGNESIUM ION, RNA (62-MER) | | Authors: | Bonneau, E, Girard, N, Lemieux, S, Legault, P. | | Deposit date: | 2015-06-09 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the II-III-VI three-way junction from the Neurospora VS ribozyme reveals a critical tertiary interaction and provides new insights into the global ribozyme structure.
Rna, 21, 2015
|
|
2N3Q
 
 | |
2LOX
 
 | |
