1MQV
 
 | | Crystal Structure of the Q1A/F32W/W72F mutant of Rhodopseudomonas palustris cytochrome c' (prime) expressed in E. coli | | Descriptor: | CYTOCHROME C', HEME C | | Authors: | Lee, J.C, Engman, K.C, Tezcan, F.A, Gray, H.B, Winkler, J.R. | | Deposit date: | 2002-09-17 | | Release date: | 2002-11-20 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Features of Cytochrome c' Folding Intermediates Revealed by Fluorescence Energy-Transfer Kinetics
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3QJN
 
 | | Structural flexibility of Shank PDZ domain is important for its binding to different ligands | | Descriptor: | Beta-PIX, SH3 and multiple ankyrin repeat domains protein 1 | | Authors: | Lee, J.H, Park, H, Park, S.J, Kim, H.J, Eom, S.H. | | Deposit date: | 2011-01-30 | | Release date: | 2011-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | The structural flexibility of the shank1 PDZ domain is important for its binding to different ligands
Biochem.Biophys.Res.Commun., 407, 2011
|
|
4MRR
 
 | | Structure of a bacterial Atm1-family ABC transporter | | Descriptor: | ABC transporter related protein, LAURYL DIMETHYLAMINE-N-OXIDE, PHOSPHATE ION, ... | | Authors: | Lee, J.Y, Yang, J.G, Zhitnitsky, D, Lewinson, O, Rees, D.C. | | Deposit date: | 2013-09-17 | | Release date: | 2014-03-19 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural basis for heavy metal detoxification by an Atm1-type ABC exporter.
Science, 343, 2014
|
|
2FB9
 
 | |
4MRV
 
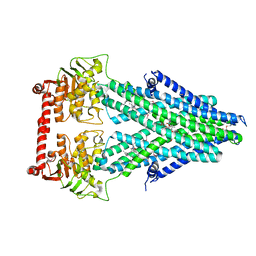 | | Structure of a bacterial Atm1-family ABC transporter | | Descriptor: | ABC transporter related protein, LAURYL DIMETHYLAMINE-N-OXIDE, PHOSPHATE ION, ... | | Authors: | Lee, J.Y, Yang, J.G, Zhitnitsky, D, Lewinson, O, Rees, D.C. | | Deposit date: | 2013-09-17 | | Release date: | 2014-03-19 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for heavy metal detoxification by an Atm1-type ABC exporter.
Science, 343, 2014
|
|
3NE2
 
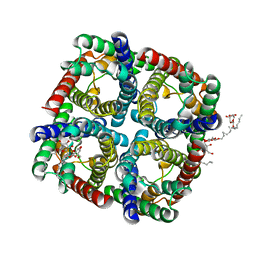 | |
4MRP
 
 | | Structure of a bacterial Atm1-family ABC transporter | | Descriptor: | ABC transporter related protein, GLUTATHIONE, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Lee, J.Y, Yang, J.G, Zhitnitsky, D, Lewinson, O, Rees, D.C. | | Deposit date: | 2013-09-17 | | Release date: | 2014-03-19 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for heavy metal detoxification by an Atm1-type ABC exporter.
Science, 343, 2014
|
|
3O3Q
 
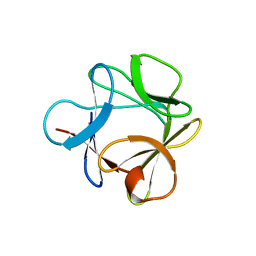 | |
3H2U
 
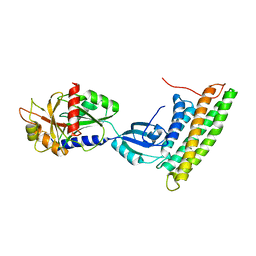 | | Human raver1 RRM1, RRM2, and RRM3 domains in complex with human vinculin tail domain Vt | | Descriptor: | Raver-1, Vinculin | | Authors: | Lee, J.H, Rangarajan, E.S, Yogesha, S.D, Izard, T. | | Deposit date: | 2009-04-14 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Raver1 interactions with vinculin and RNA suggest a feed-forward pathway in directing mRNA to focal adhesions
Structure, 17, 2009
|
|
7JW5
 
 | | Crystal structure of WT-CYP199A4 in complex with 4-phenylbenzoic acid | | Descriptor: | CHLORIDE ION, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lee, J.H.Z, Bruning, J.B, Bell, S.G. | | Deposit date: | 2020-08-24 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.526 Å) | | Cite: | Different Geometric Requirements for Cytochrome P450-Catalyzed Aliphatic Versus Aromatic Hydroxylation Results in Chemoselective Oxidation
Acs Catalysis, 12, 2022
|
|
1SD2
 
 | | STRUCTURE OF HUMAN 5'-DEOXY-5'-METHYLTHIOADENOSINE PHOSPHORYLASE COMPLEXED WITH 5'-METHYLTHIOTUBERCIDIN | | Descriptor: | 2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-METHYLSULFANYLMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, 5'-methylthioadenosine phosphorylase, SULFATE ION | | Authors: | Lee, J.E, Settembre, E.C, Cornell, K.A, Riscoe, M.K, Sufrin, J.R, Ealick, S.E, Howell, P.L. | | Deposit date: | 2004-02-12 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Comparison of MTA Phosphorylase and MTA/AdoHcy Nucleosidase Explains Substrate Preferences and Identifies Regions Exploitable for Inhibitor Design.
Biochemistry, 43, 2004
|
|
3BAQ
 
 | |
3BAO
 
 | |
3BAV
 
 | |
3BB2
 
 | |
5H3H
 
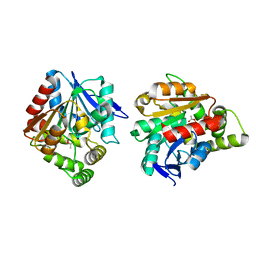 | | Esterase (EaEST) from Exiguobacterium antarcticum | | Descriptor: | Abhydrolase domain-containing protein, ETHANEPEROXOIC ACID | | Authors: | Lee, J.H, Lee, C.W. | | Deposit date: | 2016-10-24 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure and Functional Characterization of an Esterase (EaEST) from Exiguobacterium antarcticum.
Plos One, 12, 2017
|
|
1YTO
 
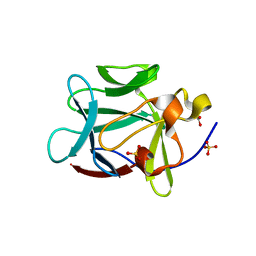 | | Crystal Structure of Gly19 deletion Mutant of Human Acidic Fibroblast Growth Factor | | Descriptor: | FORMIC ACID, Heparin-binding growth factor 1, SULFATE ION | | Authors: | Lee, J, Blaber, M. | | Deposit date: | 2005-02-10 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conversion of type I 4:6 to 3:5 beta-turn types in human acidic fibroblast growth factor: Effects upon structure, stability, folding, and mitogenic function.
Proteins, 62, 2006
|
|
4QXL
 
 | | Crystal Structure of FLHE | | Descriptor: | Flagellar protein flhE | | Authors: | Lee, J, Monzingo, A.F, Keatinge-Clay, A.T, Harshey, R.M. | | Deposit date: | 2014-07-21 | | Release date: | 2015-01-14 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.512 Å) | | Cite: | Structure of Salmonella FlhE, Conserved Member of a Flagellar Type III Secretion Operon.
J.Mol.Biol., 427, 2015
|
|
2EVU
 
 | | Crystal structure of aquaporin AqpM at 2.3A resolution | | Descriptor: | Aquaporin aqpM, GLYCEROL, octyl beta-D-glucopyranoside | | Authors: | Lee, J.K, Kozono, D, Remis, J, Kitagawa, Y, Agre, P, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2005-10-31 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for conductance by the archaeal aquaporin AqpM at 1.68 A.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1CFL
 
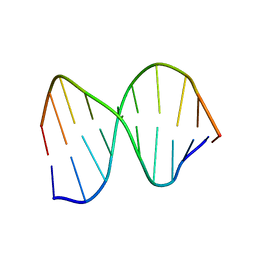 | | DNA DECAMER DUPLEX CONTAINING T5-T6 PHOTOADDUCT | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*(64T)P*TP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*TP*GP*AP*TP*GP*CP*G)-3') | | Authors: | Lee, J.-H, Hwang, G.-S, Choi, B.-S. | | Deposit date: | 1999-03-19 | | Release date: | 1999-05-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA decamer duplex containing the stable 3' T.G base pair of the pyrimidine(6-4)pyrimidone photoproduct [(6-4) adduct]: implications for the highly specific 3' T --> C transition of the (6-4) adduct.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
6KTA
 
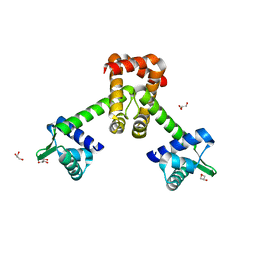 | | Crystal structure of B. halodurans MntR in apo form | | Descriptor: | GLYCEROL, HTH-type transcriptional regulator MntR | | Authors: | Lee, J.Y, Lee, M.Y. | | Deposit date: | 2019-08-26 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of the manganese transport regulator MntR from Bacillus halodurans in apo and manganese bound forms.
Plos One, 14, 2019
|
|
4MVD
 
 | | Crystal Structure of a Mammalian Cytidylyltransferase | | Descriptor: | Choline-phosphate cytidylyltransferase A, [2-CYTIDYLATE-O'-PHOSPHONYLOXYL]-ETHYL-TRIMETHYL-AMMONIUM | | Authors: | Lee, J, Cornell, R.B. | | Deposit date: | 2013-09-23 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (8 Å) | | Cite: | Structural Basis for Autoinhibition of CTP:Phosphocholine Cytidylyltransferase (CCT), the Regulatory Enzyme in Phosphatidylcholine Synthesis, by Its Membrane-binding Amphipathic Helix.
J.Biol.Chem., 289, 2014
|
|
1SD1
 
 | | STRUCTURE OF HUMAN 5'-DEOXY-5'-METHYLTHIOADENOSINE PHOSPHORYLASE COMPLEXED WITH FORMYCIN A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, 5'-methylthioadenosine phosphorylase | | Authors: | Lee, J.E, Settembre, E.C, Cornell, K.A, Riscoe, M.K, Sufrin, J.R, Ealick, S.E, Howell, P.L. | | Deposit date: | 2004-02-12 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Comparison of MTA Phosphorylase and MTA/AdoHcy Nucleosidase Explains Substrate Preferences and Identifies Regions Exploitable for Inhibitor Design.
Biochemistry, 43, 2004
|
|
1Z4S
 
 | |
1QL5
 
 | | DNA DECAMER DUPLEX CONTAINING T5-T6 PHOTOADDUCT | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*TP*+TP*AP*CP*GP*C)- 3'), DNA (5'-D(*GP*CP*GP*TP*TP*AP*TP*GP*CP*G)-3') | | Authors: | Lee, J.-H, Hwang, G.-S, Choi, B.-S. | | Deposit date: | 1999-08-24 | | Release date: | 2000-04-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a DNA Decamer Duplex Containing the 3' T.T Base Pair of the Cis-Syn Cyclobutane Pyrimidine Dimer: Implication for the Mutagenic Property of the Cis-Syn Dimer.
Nucleic Acids Res., 28, 2000
|
|
