3P6Q
 
 | |
3P6P
 
 | | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-C6-Bio | | Descriptor: | (3S,5S,7S)-N-[6-({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexyl]tricyclo[3.3.1.1~3,7~]decane-1-carboxamide, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Lee, Y.-T, Wilson, R.F, Glazer, E.C, Goodin, D.B. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-C6-Bio
To be Published
|
|
3P6W
 
 | |
3P6O
 
 | | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-Etg-Dans | | Descriptor: | (3S,5S,7S)-N-(2-{2-[2-({[5-(dimethylamino)naphthalen-1-yl]sulfonyl}amino)ethoxy]ethoxy}ethyl)tricyclo[3.3.1.1~3,7~]decane-1-carboxamide, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Lee, Y.-T, Wilson, R.F, Glazer, E.C, Goodin, D.B. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-Etg-Dans
To be Published
|
|
3P6T
 
 | | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC2-C8-Dans | | Descriptor: | Camphor 5-monooxygenase, N-[8-({[5-(dimethylamino)naphthalen-1-yl]sulfonyl}amino)octyl]-2-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]acetamide, POTASSIUM ION, ... | | Authors: | Lee, Y.-T, Wilson, R.F, Glazer, E.C, Goodin, D.B. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC2-C8-Dans
To be Published
|
|
2JTT
 
 | |
3P6R
 
 | |
3P6U
 
 | |
3P6X
 
 | | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC3-C8-Dans | | Descriptor: | Camphor 5-monooxygenase, N-[8-({[5-(dimethylamino)naphthalen-1-yl]sulfonyl}amino)octyl]-3-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]propanamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lee, Y.-T, Wilson, R.F, Glazer, E.C, Goodin, D.B. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC3-C8-Dans
To be Published
|
|
3P6S
 
 | |
3P6V
 
 | |
6AGO
 
 | | Crystal structure of MRG15-ASH1L Histone methyltransferase complex | | Descriptor: | Histone-lysine N-methyltransferase ASH1L, Mortality factor 4 like 1, S-ADENOSYLMETHIONINE, ... | | Authors: | Lee, Y, Song, J. | | Deposit date: | 2018-08-13 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Structural Basis of MRG15-Mediated Activation of the ASH1L Histone Methyltransferase by Releasing an Autoinhibitory Loop.
Structure, 27, 2019
|
|
5EUR
 
 | | Hypothetical protein SF216 from shigella flexneri 5a M90T | | Descriptor: | MAGNESIUM ION, SULFATE ION, Uncharacterized protein | | Authors: | Lee, Y.-S, Seok, S.-H, Kim, H.-N, An, J.-G, Chung, K.Y, Won, H.-S, Seo, M.-D. | | Deposit date: | 2015-11-19 | | Release date: | 2016-11-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Crystal structure of SF216 from Shigella flexneri strain 5a
To Be Published
|
|
6JMR
 
 | | CD98hc extracellular domain bound to HBJ127 Fab and MEM-108 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, ... | | Authors: | Lee, Y, Nishizawa, T, Kusakizako, T, Oda, K, Ishitani, R, Yokoyama, T, Nakane, T, Shirouzu, M, Nureki, O. | | Deposit date: | 2019-03-13 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the human L-type amino acid transporter 1 in complex with glycoprotein CD98hc.
Nat.Struct.Mol.Biol., 26, 2019
|
|
2F7N
 
 | | Structure of D. radiodurans Dps-1 | | Descriptor: | COBALT (II) ION, DNA-binding stress response protein, Dps family, ... | | Authors: | Lee, Y.H, Kim, S.G, Bhattacharyya, G, Grove, A. | | Deposit date: | 2005-12-01 | | Release date: | 2006-11-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Dps-1, a functionally distinct Dps protein from Deinococcus radiodurans.
J.Mol.Biol., 361, 2006
|
|
6JMQ
 
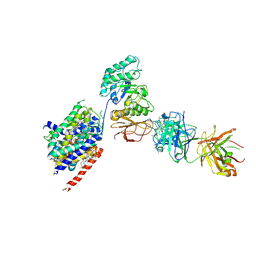 | | LAT1-CD98hc complex bound to MEM-108 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, ... | | Authors: | Lee, Y, Nishizawa, T, Kusakizako, T, Oda, K, Ishitani, R, Nakane, T, Nureki, O. | | Deposit date: | 2019-03-13 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Cryo-EM structure of the human L-type amino acid transporter 1 in complex with glycoprotein CD98hc.
Nat.Struct.Mol.Biol., 26, 2019
|
|
5Y78
 
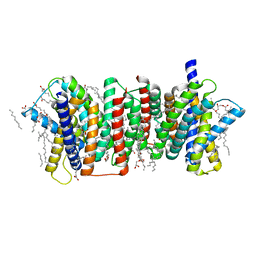 | | Crystal structure of the triose-phosphate/phosphate translocator in complex with inorganic phosphate | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, PHOSPHATE ION, Putative hexose phosphate translocator | | Authors: | Lee, Y, Nishizawa, T, Takemoto, M, Kumazaki, K, Yamashita, K, Hirata, K, Minoda, A, Nagatoishi, S, Tsumoto, K, Ishitani, R, Nureki, O. | | Deposit date: | 2017-08-16 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the triose-phosphate/phosphate translocator reveals the basis of substrate specificity
Nat Plants, 3, 2017
|
|
5Y79
 
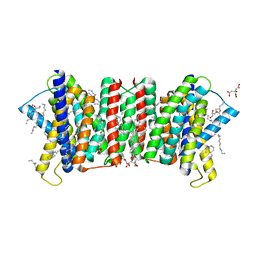 | | Crystal structure of the triose-phosphate/phosphate translocator in complex with 3-phosphoglycerate | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-PHOSPHOGLYCERIC ACID, CITRATE ANION, ... | | Authors: | Lee, Y, Nishizawa, T, Takemoto, M, Kumazaki, K, Yamashita, K, Hirata, K, Minoda, A, Nagatoishi, S, Tsumoto, K, Ishitani, R, Nureki, O. | | Deposit date: | 2017-08-16 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the triose-phosphate/phosphate translocator reveals the basis of substrate specificity
Nat Plants, 3, 2017
|
|
6JG0
 
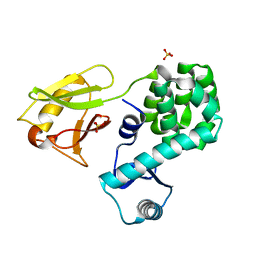 | | Crystal structure of the N-terminal domain single mutant (S92E) of the human mitochondrial calcium uniporter fused with T4 lysozyme | | Descriptor: | Endolysin,Calcium uniporter protein, SULFATE ION | | Authors: | Lee, Y, Park, J, Min, C.K, Kang, J.Y, Kim, T.G, Yamamoto, T, Kim, D.H, Eom, S.H. | | Deposit date: | 2019-02-13 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of calcium channel domain
To Be Published
|
|
4Y1L
 
 | | Ubc9 Homodimer The Missing Link in Poly-SUMO Chain Formation | | Descriptor: | RWD domain-containing protein 3, SUMO-conjugating enzyme UBC9 | | Authors: | Aileen, Y.A, Ambaye, N.D, Li, Y.J, Vega, R, Bzymek, K, Williams, J.C, Hu, W, Chen, Y. | | Deposit date: | 2015-02-08 | | Release date: | 2015-05-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | RWD Domain as an E2 (Ubc9)-Interaction Module.
J.Biol.Chem., 290, 2015
|
|
4MM2
 
 | | Crystal structure of yeast primase catalytic subunit | | Descriptor: | CADMIUM ION, CITRIC ACID, DNA primase small subunit | | Authors: | Park, K.R, An, J.Y, Lee, Y, Youn, H.S, Lee, J.G, Kang, J.Y, Kim, T.G, Lim, J.J, Eom, S.H, Wang, J. | | Deposit date: | 2013-09-07 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of yeast primase catalytic subunit
To be Published
|
|
4R1P
 
 | |
7XM8
 
 | | Glucagon amyloid fibril | | Descriptor: | Glucagon | | Authors: | Jeong, H, Lin, Y, Lee, Y.-H. | | Deposit date: | 2022-04-25 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomistic zipper-like amyloid structure of full-length glucagon
To Be Published
|
|
2A3P
 
 | | Structure of Desulfovibrio desulfuricans G20 tetraheme cytochrome with bound molybdate | | Descriptor: | COG3005: Nitrate/TMAO reductases, membrane-bound tetraheme cytochrome c subunit, HEME C, ... | | Authors: | Pattarkine, M.V, Lee, Y.-H, Tanner, J.J, Wall, J.D. | | Deposit date: | 2005-06-25 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Desulfovibrio desulfuricans G20 Tetraheme Cytochrome Structure at 1.5A and Cytochrome Interaction with Metal Complexes
J.Mol.Biol., 358, 2006
|
|
2A3M
 
 | | Structure of Desulfovibrio desulfuricans G20 tetraheme cytochrome (oxidized form) | | Descriptor: | COG3005: Nitrate/TMAO reductases, membrane-bound tetraheme cytochrome c subunit, HEME C | | Authors: | Pattarkine, M.V, Tanner, J.J, Bottoms, C.A, Lee, Y.H, Wall, J.D. | | Deposit date: | 2005-06-25 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Desulfovibrio desulfuricans G20 Tetraheme Cytochrome Structure at 1.5A and Cytochrome Interaction with Metal Complexes
J.Mol.Biol., 358, 2006
|
|
