2A5A
 
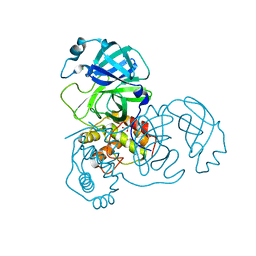 | | Crystal structure of unbound SARS coronavirus main peptidase in the space group C2 | | Descriptor: | 1,2-ETHANEDIOL, 3C-like peptidase, CHLORIDE ION | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N. | | Deposit date: | 2005-06-30 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal Structures of the Main Peptidase from the SARS Coronavirus Inhibited by a Substrate-like Aza-peptide Epoxide
J.Mol.Biol., 353, 2005
|
|
4E8Y
 
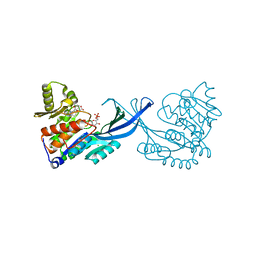 | | Crystal Structure of Burkholderia cenocepacia HldA in Complex with an ATP-competitive Inhibitor | | Descriptor: | 7-O-phosphono-D-glycero-beta-D-manno-heptopyranose, CHLORIDE ION, D-beta-D-heptose 7-phosphate kinase, ... | | Authors: | Lee, T.-W, Verhey, T.B, Junop, M.S. | | Deposit date: | 2012-03-20 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural-functional studies of Burkholderia cenocepacia D-glycero-beta-D-manno-heptose 7-phosphate kinase (HldA) and characterization of inhibitors with antibiotic adjuvant and antivirulence properties.
J.Med.Chem., 56, 2013
|
|
4E8Z
 
 | | Crystal Structure of Burkholderia cenocepacia HldA in Complex with an ATP-competitive Inhibitor | | Descriptor: | D-beta-D-heptose 7-phosphate kinase, POTASSIUM ION, {[2-({[5-(2,6-dichlorophenyl)-1,2,4-triazin-3-yl]amino}methyl)-1,3-benzothiazol-5-yl]oxy}acetic acid | | Authors: | Lee, T.-W, Verhey, T.B, Junop, M.S. | | Deposit date: | 2012-03-20 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural-functional studies of Burkholderia cenocepacia D-glycero-beta-D-manno-heptose 7-phosphate kinase (HldA) and characterization of inhibitors with antibiotic adjuvant and antivirulence properties.
J.Med.Chem., 56, 2013
|
|
4E8W
 
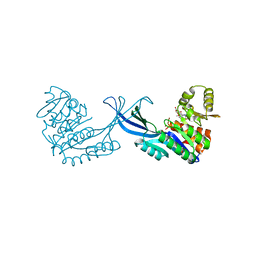 | | Crystal Structure of Burkholderia cenocepacia HldA in Complex with an ATP-competitive Inhibitor | | Descriptor: | D-beta-D-heptose 7-phosphate kinase, POTASSIUM ION, {[2-({[5-(2,6-dimethoxyphenyl)-1,2,4-triazin-3-yl]amino}methyl)-1,3-benzothiazol-5-yl]oxy}acetic acid | | Authors: | Lee, T.-W, Verhey, T.B, Junop, M.S. | | Deposit date: | 2012-03-20 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8654 Å) | | Cite: | Structural-functional studies of Burkholderia cenocepacia D-glycero-beta-D-manno-heptose 7-phosphate kinase (HldA) and characterization of inhibitors with antibiotic adjuvant and antivirulence properties.
J.Med.Chem., 56, 2013
|
|
4E84
 
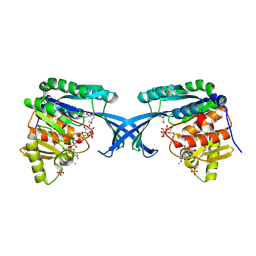 | | Crystal Structure of Burkholderia cenocepacia HldA | | Descriptor: | 1,7-di-O-phosphono-D-glycero-beta-D-manno-heptopyranose, 7-O-phosphono-D-glycero-beta-D-manno-heptopyranose, CHLORIDE ION, ... | | Authors: | Lee, T.-W, Junop, M.S. | | Deposit date: | 2012-03-19 | | Release date: | 2012-12-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural-functional studies of Burkholderia cenocepacia D-glycero-beta-D-manno-heptose 7-phosphate kinase (HldA) and characterization of inhibitors with antibiotic adjuvant and antivirulence properties.
J.Med.Chem., 56, 2013
|
|
2GT8
 
 | | Crystal structure of SARS coronavirus main peptidase (with an additional Ala at the N-terminus of each protomer) in the space group P43212 | | Descriptor: | 3C-like proteinase | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N.G. | | Deposit date: | 2006-04-27 | | Release date: | 2006-12-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures Reveal an Induced-fit Binding of a Substrate-like Aza-peptide Epoxide to SARS Coronavirus Main Peptidase.
J.Mol.Biol., 366, 2007
|
|
2GT7
 
 | | Crystal structure of SARS coronavirus main peptidase at pH 6.0 in the space group P21 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3C-like proteinase | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N.G. | | Deposit date: | 2006-04-27 | | Release date: | 2006-12-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structures Reveal an Induced-fit Binding of a Substrate-like Aza-peptide Epoxide to SARS Coronavirus Main Peptidase.
J.Mol.Biol., 366, 2007
|
|
2GTB
 
 | | Crystal structure of SARS coronavirus main peptidase (with an additional Ala at the N-terminus of each protomer) inhibited by an aza-peptide epoxide in the space group P43212 | | Descriptor: | (5S,8S,14R)-ETHYL 11-(3-AMINO-3-OXOPROPYL)-8-BENZYL-14-HYDROXY-5-ISOBUTYL-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,11-TETRAAZAPENTADECAN-15-OATE, 3C-like proteinase, ACETIC ACID | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C. | | Deposit date: | 2006-04-27 | | Release date: | 2006-12-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures Reveal an Induced-fit Binding of a Substrate-like Aza-peptide Epoxide to SARS Coronavirus Main Peptidase.
J.Mol.Biol., 366, 2007
|
|
2A5I
 
 | | Crystal structures of SARS coronavirus main peptidase inhibited by an aza-peptide epoxide in the space group C2 | | Descriptor: | (5S,8S,14R)-ETHYL 11-(3-AMINO-3-OXOPROPYL)-8-BENZYL-14-HYDROXY-5-ISOBUTYL-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,11-TETRAAZAPENTADECAN-15-OATE, 1,2-ETHANEDIOL, 3C-like peptidase, ... | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N. | | Deposit date: | 2005-06-30 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structures of the Main Peptidase from the SARS Coronavirus Inhibited by a Substrate-like Aza-peptide Epoxide
J.Mol.Biol., 353, 2005
|
|
2A5K
 
 | | Crystal structures of SARS coronavirus main peptidase inhibited by an aza-peptide epoxide in space group P212121 | | Descriptor: | (5S,8S,14R)-ETHYL 11-(3-AMINO-3-OXOPROPYL)-8-BENZYL-14-HYDROXY-5-ISOBUTYL-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,11-TETRAAZAPENTADECAN-15-OATE, 3C-like peptidase | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N. | | Deposit date: | 2005-06-30 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the Main Peptidase from the SARS Coronavirus Inhibited by a Substrate-like Aza-peptide Epoxide
J.Mol.Biol., 353, 2005
|
|
4DRW
 
 | | Crystal Structure of the Ternary Complex between S100A10, an Annexin A2 N-terminal Peptide and an AHNAK Peptide | | Descriptor: | Neuroblast differentiation-associated protein AHNAK, Protein S100-A10/Annexin A2 chimeric protein | | Authors: | Rezvanpour, A, Lee, T.-W, Junop, M.S, Shaw, G.S. | | Deposit date: | 2012-02-17 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of an asymmetric ternary protein complex provides insight for membrane interaction.
Structure, 20, 2012
|
|
