2AZ0
 
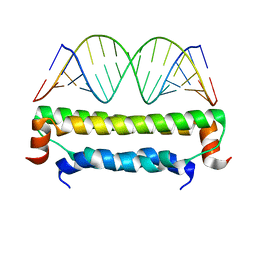 | | Flock House virus B2-dsRNA Complex (P212121) | | Descriptor: | 5'-R(*GP*CP*AP*(5BU)P*GP*GP*AP*CP*GP*CP*GP*(5BU)P*CP*CP*AP*(5BU)P*GP*C)-3', B2 protein | | Authors: | Chao, J.A, Lee, J.H, Chapados, B.R, Debler, E.W, Schneemann, A, Williamson, J.R. | | Deposit date: | 2005-09-09 | | Release date: | 2005-10-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dual modes of RNA-silencing suppression by Flock House virus protein B2.
Nat.Struct.Mol.Biol., 12, 2005
|
|
7EHK
 
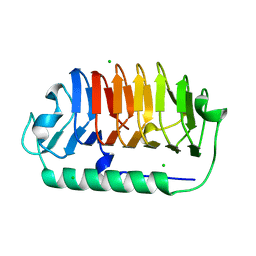 | | Crystal structure of C107S mutant of FfIBP | | Descriptor: | CHLORIDE ION, Ice-binding protein | | Authors: | Do, H, Lee, J.H. | | Deposit date: | 2021-03-29 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Importance of rigidity of ice-binding protein (FfIBP) for hyperthermal hysteresis activity and microbial survival.
Int.J.Biol.Macromol., 204, 2022
|
|
6IL9
 
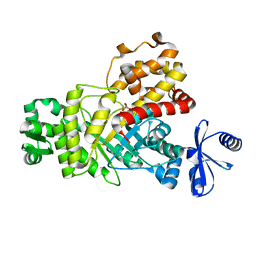 | | One Glycerol complexed Crystal structure of fructuronate-tagaturonate epimerase UxaE from Cohnella laeviribosi | | Descriptor: | Fructuronate-tagaturonate epimerase UxaE from Cohnella laeviribosi in complex with 1 glycerol, GLYCEROL, ZINC ION | | Authors: | Choi, M.Y, Kang, L.W, Ho, T.H, Nguyen, D.Q, Lee, I.H, Lee, J.H, Park, Y.S, Park, H.J. | | Deposit date: | 2018-10-17 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.72005355 Å) | | Cite: | Crystal structure of fructuronate-tagaturonate epimerase UxaE from Cohnella laeviribosi
To Be Published
|
|
3BVE
 
 | | Structural basis for the iron uptake mechanism of Helicobacter pylori ferritin | | Descriptor: | Ferritin, GLYCEROL | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Shin, H.J, Yang, I.S. | | Deposit date: | 2008-01-07 | | Release date: | 2009-01-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the iron uptake mechanism of Helicobacter pylori ferritin
To be Published
|
|
6JZL
 
 | |
6ILB
 
 | | Native crystal structure of fructuronate-tagaturonate epimerase UxaE from Cohnella laeviribosi | | Descriptor: | 1,2-ETHANEDIOL, Fructuronate-tagaturonate epimerase UxaE from Cohnella laeviribosi in complex with 1 glycerol, ZINC ION | | Authors: | Choi, M.Y, Kang, L.W, Ho, T.H, Nguyen, D.Q, Lee, I.H, Lee, J.H, Park, Y.S, Park, H.J. | | Deposit date: | 2018-10-17 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.50541973 Å) | | Cite: | Crystal structure of fructuronate-tagaturonate epimerase UxaE from Cohnella laeviribosi
To be published
|
|
3A4C
 
 | | Crystal structure of cdt1 C terminal domain | | Descriptor: | DNA replication factor Cdt1 | | Authors: | Cho, Y, Lee, J.H. | | Deposit date: | 2009-07-06 | | Release date: | 2009-10-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Structure of the Cdt1 C-terminal domain: Conservation of the winged helix fold in replication licensing factors
Protein Sci., 18, 2009
|
|
1N7F
 
 | | Crystal structure of the sixth PDZ domain of GRIP1 in complex with liprin C-terminal peptide | | Descriptor: | 8-mer peptide from interacting protein (liprin), AMPA receptor interacting protein GRIP | | Authors: | Im, Y.J, Park, S.H, Rho, S.H, Lee, J.H, Kang, G.B, Sheng, M, Kim, E, Eom, S.H. | | Deposit date: | 2002-11-14 | | Release date: | 2003-08-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of GRIP1 PDZ6-peptide complex reveals the structural basis for class II PDZ target recognition and PDZ domain-mediated multimerization
J.BIOL.CHEM., 278, 2003
|
|
1Q3P
 
 | | Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization | | Descriptor: | C-terminal hexapeptide from Guanylate kinase-associated protein, Shank1 | | Authors: | Im, Y.J, Lee, J.H, Park, S.H, Park, S.J, Rho, S.-H, Kang, G.B, Kim, E, Eom, S.H. | | Deposit date: | 2003-07-31 | | Release date: | 2004-01-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization
J.Biol.Chem., 278, 2003
|
|
6L8P
 
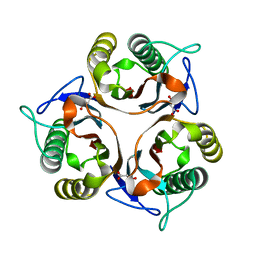 | | Crystal structure of RidA from Antarctic bacterium Psychrobacter sp. PAMC 21119 | | Descriptor: | MALONATE ION, RidA family protein | | Authors: | Kwon, S, Lee, C.W, Koh, H.Y, Lee, J.H, Park, H.H. | | Deposit date: | 2019-11-06 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Crystal structure of the reactive intermediate/imine deaminase A homolog from the Antarctic bacterium Psychrobacter sp. PAMC 21119.
Biochem.Biophys.Res.Commun., 522, 2020
|
|
6ILA
 
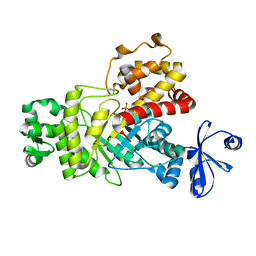 | | Two Glycerol complexed Crystal structure of fructuronate-tagaturonate epimerase UxaE from Cohnella laeviribosi | | Descriptor: | Fructuronate-tagaturonate epimerase UxaE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Choi, M.Y, Kang, L.W, Ho, T.H, Nguyen, D.Q, Lee, I.H, Lee, J.H, Park, Y.S, Park, H.J. | | Deposit date: | 2018-10-17 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of fructuronate-tagaturonate epimerase UxaE from Cohnella laeviribosi
To be published
|
|
1N7E
 
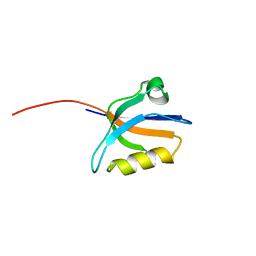 | | Crystal structure of the sixth PDZ domain of GRIP1 | | Descriptor: | AMPA receptor interacting protein GRIP | | Authors: | Im, Y.J, Park, S.H, Rho, S.H, Lee, J.H, Kang, G.B, Sheng, M, Kim, E, Eom, S.H. | | Deposit date: | 2002-11-14 | | Release date: | 2003-08-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of GRIP1 PDZ6-peptide complex reveals the structural basis for class II PDZ target recognition and PDZ domain-mediated multimerization
J.BIOL.CHEM., 278, 2003
|
|
1Q3O
 
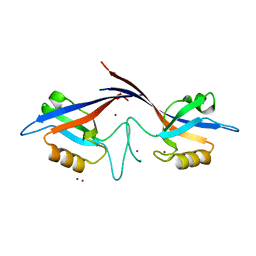 | | Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization | | Descriptor: | BROMIDE ION, Shank1 | | Authors: | Im, Y.J, Lee, J.H, Park, S.H, Park, S.J, Rho, S.-H, Kang, G.B, Kim, E, Eom, S.H. | | Deposit date: | 2003-07-31 | | Release date: | 2004-01-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization
J.Biol.Chem., 278, 2003
|
|
1XHK
 
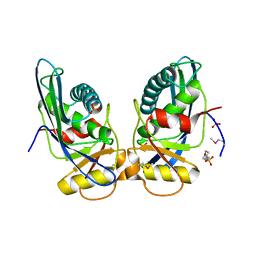 | | Crystal structure of M. jannaschii Lon proteolytic domain | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative protease La homolog, SULFATE ION | | Authors: | Im, Y.J, Na, Y, Kang, G.B, Rho, S.-H, Kim, M.-K, Lee, J.H, Chung, C.H, Eom, S.H. | | Deposit date: | 2004-09-20 | | Release date: | 2004-10-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The active site of a lon protease from Methanococcus jannaschii distinctly differs from the canonical catalytic Dyad of Lon proteases.
J.Biol.Chem., 279, 2004
|
|
1YNX
 
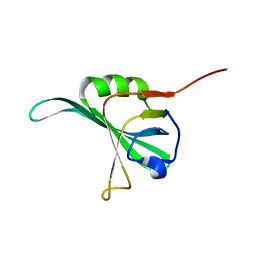 | |
7E8N
 
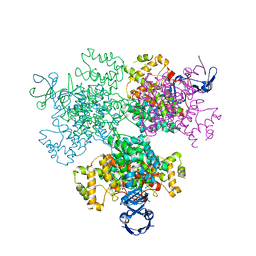 | | Crystal structure of Type II citrate synthase (HyCS) from Hymenobacter sp. PAMC 26554 | | Descriptor: | CITRIC ACID, Citrate synthase | | Authors: | Park, S.-H, Lee, C.W, Bae, D.-W, Lee, J.H. | | Deposit date: | 2021-03-02 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the cooperative activation of type II citrate synthase (HyCS) from Hymenobacter sp. PAMC 26554.
Int.J.Biol.Macromol., 183, 2021
|
|
2O4C
 
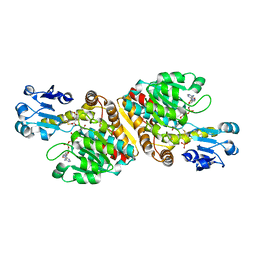 | | Crystal Structure of D-Erythronate-4-phosphate Dehydrogenase Complexed with NAD | | Descriptor: | Erythronate-4-phosphate dehydrogenase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Ha, J.Y, Lee, J.H, Kim, K.H, Kim, D.J, Lee, H.H, Kim, H.K, Yoon, H.J, Suh, S.W. | | Deposit date: | 2006-12-04 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of d-Erythronate-4-phosphate Dehydrogenase Complexed with NAD
J.Mol.Biol., 366, 2007
|
|
1XNH
 
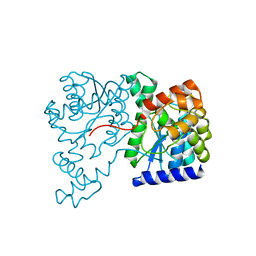 | | Crystal Structure of NH3-dependent NAD+ synthetase from Helicobacter pylori | | Descriptor: | NH(3)-dependent NAD(+) synthetase | | Authors: | Kang, G.B, Kim, Y.S, Im, Y.J, Rho, S.H, Lee, J.H, Eom, S.H. | | Deposit date: | 2004-10-05 | | Release date: | 2005-04-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of NH3-dependent NAD+ synthetase from Helicobacter pylori
Proteins, 58, 2005
|
|
1XNG
 
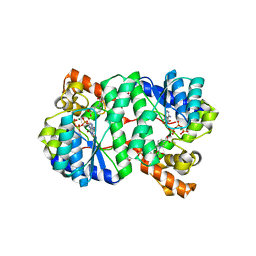 | | Crystal Structure of NH3-dependent NAD+ synthetase from Helicobacter pylori | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, NH(3)-dependent NAD(+) synthetase, ... | | Authors: | Kang, G.B, Kim, Y.S, Im, Y.J, Rho, S.H, Lee, J.H, Eom, S.H. | | Deposit date: | 2004-10-05 | | Release date: | 2005-04-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of NH3-dependent NAD+ synthetase from Helicobacter pylori
Proteins, 58, 2005
|
|
6JKW
 
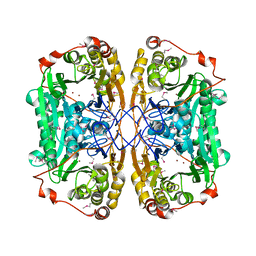 | | Seleno-methionine PNGM-1 from deep-sea sediment metagenome | | Descriptor: | Metallo-beta-lactamases PNGM-1, ZINC ION | | Authors: | Hong, M.K, Park, K.S, Jeon, J.H, Lee, J.H, Park, Y.S, Lee, S.H, Kang, L.W. | | Deposit date: | 2019-03-02 | | Release date: | 2019-04-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | PNGM 1 a novel subclass B3 metallo beta lactamase from a deep sea sediment metagenome
Journal of Global Antimicrobial Resistance, 14, 2018
|
|
3TJ6
 
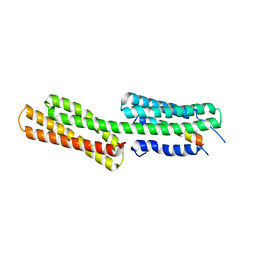 | | human vinculin head domain (Vh1, residues 1-258) in complex with the vinculin binding site of the surface cell antigen 4 (sca4-VBS-C; residues 812-835) from Rickettsia rickettsii | | Descriptor: | Antigenic heat-stable 120 kDa protein, Vinculin | | Authors: | Park, H, Lee, J.H, Gouin, E, Cossart, P, Izard, T. | | Deposit date: | 2011-08-23 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | The rickettsia surface cell antigen 4 applies mimicry to bind to and activate vinculin.
J.Biol.Chem., 286, 2011
|
|
3TJ5
 
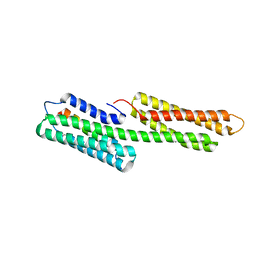 | | human vinculin head domain (Vh1, residues 1-258) in complex with the vinculin binding site of the surface cell antigen 4 (sca4-VBS-N; residues 412-434) from Rickettsia rickettsii | | Descriptor: | Antigenic heat-stable 120 kDa protein, GLYCEROL, Vinculin | | Authors: | Park, H, Lee, J.H, Gouin, E, Cossart, P, Izard, T. | | Deposit date: | 2011-08-23 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The rickettsia surface cell antigen 4 applies mimicry to bind to and activate vinculin.
J.Biol.Chem., 286, 2011
|
|
3QID
 
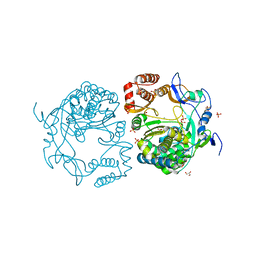 | | Crystal structures and functional analysis of murine norovirus RNA-dependent RNA polymerase | | Descriptor: | GLYCEROL, MANGANESE (III) ION, RNA dependent RNA polymerase, ... | | Authors: | Kim, K.H, Intekhab, A, Lee, J.H. | | Deposit date: | 2011-01-27 | | Release date: | 2011-12-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of murine norovirus-1 RNA-dependent RNA polymerase.
J.Gen.Virol., 92, 2011
|
|
3SMZ
 
 | |
6K84
 
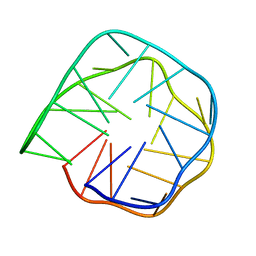 | | Structure of anti-prion RNA aptamer | | Descriptor: | RNA (25-MER) | | Authors: | Mashima, T, Lee, J.H, Hayashi, T, Nagata, T, Kinoshita, M, Katahira, M. | | Deposit date: | 2019-06-11 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Development and structural determination of an anti-PrPCaptamer that blocks pathological conformational conversion of prion protein.
Sci Rep, 10, 2020
|
|
