6JN1
 
 | | Structure of H247A mutant peptidoglycan peptidase complex with penta peptide | | 分子名称: | C0O-DAL-DAL, Peptidase M23, ZINC ION | | 著者 | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | 登録日 | 2019-03-13 | | 公開日 | 2020-01-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.382 Å) | | 主引用文献 | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN0
 
 | | Structure of H247A mutant peptidoglycan peptidase complex with tetra-tri peptide | | 分子名称: | C0O-DAL-API, Peptidase M23, ZINC ION | | 著者 | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | 登録日 | 2019-03-13 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.164 Å) | | 主引用文献 | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN7
 
 | | Structure of H216A mutant closed form peptidoglycan peptidase | | 分子名称: | D(-)-TARTARIC ACID, Peptidase M23, ZINC ION | | 著者 | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | 登録日 | 2019-03-13 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JMY
 
 | | Structure of wild type closed form of peptidoglycan peptidase | | 分子名称: | CITRIC ACID, Peptidase M23, ZINC ION | | 著者 | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | 登録日 | 2019-03-13 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.661 Å) | | 主引用文献 | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
7VOV
 
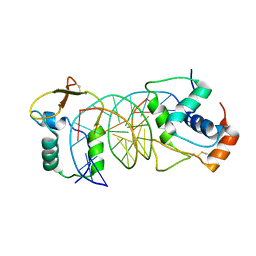 | |
7VOX
 
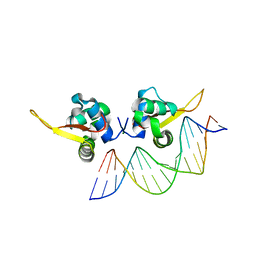 | | The crystal structure of human forkhead box protein A in complex with DNA 2 | | 分子名称: | DNA (5'-D(P*AP*AP*AP*TP*AP*TP*TP*TP*AP*TP*TP*AP*TP*CP*GP*A)-3'), DNA (5'-D(P*TP*CP*GP*AP*TP*AP*AP*TP*AP*AP*AP*TP*AP*TP*TP*T)-3'), Hepatocyte nuclear factor 3-alpha, ... | | 著者 | Choi, Y, Yoon, H.J, Lee, H.H. | | 登録日 | 2021-10-15 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | FOXL2 and FOXA1 cooperatively assemble on the TP53 promoter in alternative dimer configurations.
Nucleic Acids Res., 50, 2022
|
|
6KV1
 
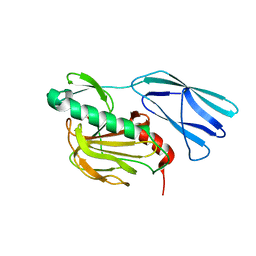 | | Structure of wild type closed form of peptidoglycan peptidase ZN SAD | | 分子名称: | CITRIC ACID, Peptidase M23, ZINC ION | | 著者 | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | 登録日 | 2019-09-03 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.722 Å) | | 主引用文献 | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
7E64
 
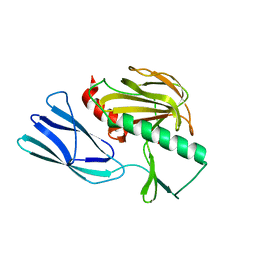 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 2-2 | | 分子名称: | 2-[[(3S)-3-acetamido-4-[[(2R)-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]amino]-4-oxidanylidene-butyl]amino]ethanoic acid, Peptidase M23, ZINC ION | | 著者 | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | 登録日 | 2021-02-21 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E67
 
 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 3-2 | | 分子名称: | N-oxidanyl-2-[4-(4-sulfamoylphenyl)phenyl]ethanamide, Peptidase M23, ZINC ION | | 著者 | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | 登録日 | 2021-02-21 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E66
 
 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 3-1 | | 分子名称: | N-[2-(oxidanylamino)-2-oxidanylidene-ethyl]-2-(4-sulfamoylphenyl)ethanamide, Peptidase M23, ZINC ION | | 著者 | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | 登録日 | 2021-02-21 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
5YU4
 
 | | Structural basis for recognition of L-lysine, L-ornithine, and L-2,4-diamino butyric acid by lysine cyclodeaminase | | 分子名称: | 2,4-DIAMINOBUTYRIC ACID, Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Min, K.J, Yoon, H.J, Matsuura, A, Kim, Y.H, Lee, H.H. | | 登録日 | 2017-11-20 | | 公開日 | 2018-05-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.144 Å) | | 主引用文献 | Structural Basis for Recognition of L-lysine, L-ornithine, and L-2,4-diamino Butyric Acid by Lysine Cyclodeaminase.
Mol. Cells, 41, 2018
|
|
5Z2W
 
 | | Crystal structure of the bacterial cell division protein FtsQ and FtsB | | 分子名称: | Cell division protein FtsB, Cell division protein FtsQ, MAGNESIUM ION | | 著者 | Choi, Y, Yoon, H.J, Lee, H.H. | | 登録日 | 2018-01-04 | | 公開日 | 2019-01-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Insights into the FtsQ/FtsB/FtsL Complex, a Key Component of the Divisome.
Sci Rep, 8, 2018
|
|
5YU3
 
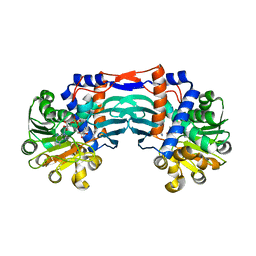 | | Structural basis for recognition of L-lysine, L-ornithine, and L-2,4-diamino butyric acid by lysine cyclodeaminase | | 分子名称: | Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROLINE, ... | | 著者 | Min, K.J, Yoon, H.J, Matsuura, A, Kim, Y.H, Lee, H.H. | | 登録日 | 2017-11-20 | | 公開日 | 2018-05-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Structural Basis for Recognition of L-lysine, L-ornithine, and L-2,4-diamino Butyric Acid by Lysine Cyclodeaminase.
Mol. Cells, 41, 2018
|
|
5YU0
 
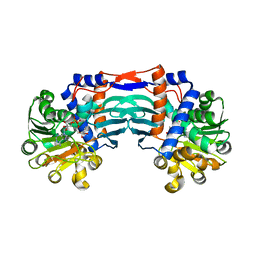 | | Structural basis for recognition of L-lysine, L-ornithine, and L-2,4-diamino butyric acid by lysine cyclodeaminase | | 分子名称: | Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | 著者 | Min, K.J, Yoon, H.J, Matsuura, A, Kim, Y.H, Lee, H.H. | | 登録日 | 2017-11-20 | | 公開日 | 2018-05-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structural Basis for Recognition of L-lysine, L-ornithine, and L-2,4-diamino Butyric Acid by Lysine Cyclodeaminase.
Mol. Cells, 41, 2018
|
|
5YU1
 
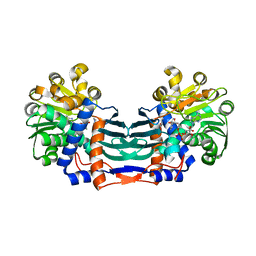 | | Structural basis for recognition of L-lysine, L-ornithine, and L-2,4-diamino butyric acid by lysine cyclodeaminase | | 分子名称: | (2S)-piperidine-2-carboxylic acid, Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Min, K.J, Yoon, H.J, Matsuura, A, Kim, Y.H, Lee, H.H. | | 登録日 | 2017-11-20 | | 公開日 | 2018-05-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.923 Å) | | 主引用文献 | Structural Basis for Recognition of L-lysine, L-ornithine, and L-2,4-diamino Butyric Acid by Lysine Cyclodeaminase.
Mol. Cells, 41, 2018
|
|
7E65
 
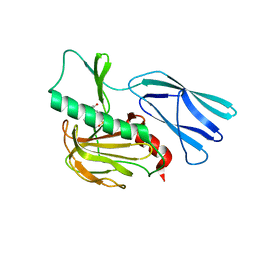 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 3 | | 分子名称: | (2S)-2-acetamido-N-[(2R)-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]-3-(4-sulfamoylphenyl)propanamide, Peptidase M23, ZINC ION | | 著者 | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | 登録日 | 2021-02-21 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E60
 
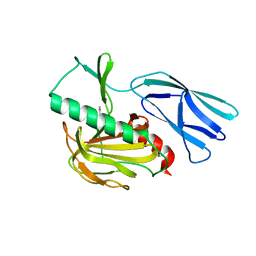 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 1 | | 分子名称: | (2~{R},6~{S})-2,6-diacetamido-7-[[(2~{R})-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]amino]-7-oxidanylidene-heptanoic acid, Peptidase M23, ZINC ION | | 著者 | Min, K, Yoon, H.J, Choi, Y, Lee, H.H. | | 登録日 | 2021-02-21 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E69
 
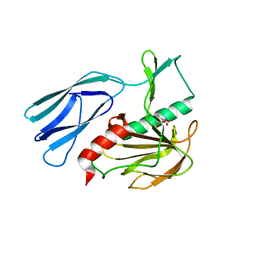 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 3-3 | | 分子名称: | N-oxidanyl-4-[(4-sulfamoylphenyl)methyl]benzamide, Peptidase M23, ZINC ION | | 著者 | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | 登録日 | 2021-02-21 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E61
 
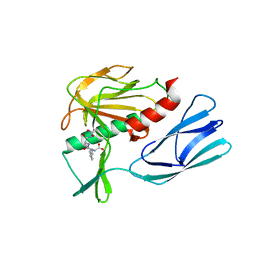 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 2 | | 分子名称: | 2-[[(3S)-3-acetamido-4-[[(2R)-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]amino]-4-oxidanylidene-butyl]-(phenylmethyl)amino]ethanoic acid, Peptidase M23, ZINC ION | | 著者 | Min, K.J, Yoon, H.J, Choi, Y, Lee, H.H. | | 登録日 | 2021-02-21 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E63
 
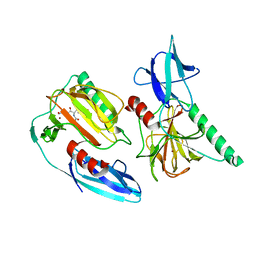 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 2-1 | | 分子名称: | 2-[[(3S)-3-acetamido-4-[[(2R)-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]amino]-4-oxidanylidene-butyl]-(cyclopentylmethyl)amino]ethanoic acid, Peptidase M23, ZINC ION | | 著者 | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | 登録日 | 2021-02-21 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7X6I
 
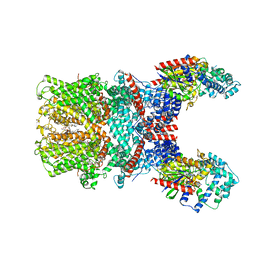 | | Cryo-EM structure of the human TRPC5 ion channel in complex with G alpha i3 subunits, class1 | | 分子名称: | (2S)-2-(hexadecanoyloxy)-3-hydroxypropyl (9Z)-octadec-9-enoate, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | 著者 | Won, J, Jeong, H, Lee, H.H. | | 登録日 | 2022-03-07 | | 公開日 | 2023-04-26 | | 最終更新日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (3.93 Å) | | 主引用文献 | Molecular architecture of the G alpha i -bound TRPC5 ion channel.
Nat Commun, 14, 2023
|
|
7X6C
 
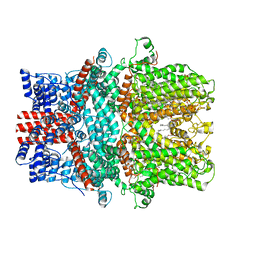 | | Cryo-EM structure of the human TRPC5 ion channel in lipid nanodiscs, class1 | | 分子名称: | (2S)-2-(hexadecanoyloxy)-3-hydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CALCIUM ION, ... | | 著者 | Won, J, Jeong, H, Lee, H.H. | | 登録日 | 2022-03-07 | | 公開日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Molecular architecture of the G alpha i -bound TRPC5 ion channel.
Nat Commun, 14, 2023
|
|
7XQW
 
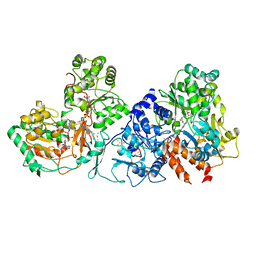 | | Formate dehydrogenase (FDH) from Methylobacterium extorquens AM1 (MeFDH1) | | 分子名称: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | 著者 | Park, J, Heo, Y.Y, Roh, S.H, Lee, H.H. | | 登録日 | 2022-05-09 | | 公開日 | 2023-08-09 | | 実験手法 | ELECTRON MICROSCOPY (2.83 Å) | | 主引用文献 | Enzymatic conversion of CO2 in real flue gas to molar-scale formate
To Be Published
|
|
6JJL
 
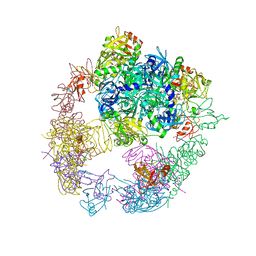 | | Crystal structure of the DegP dodecamer with a modulator | | 分子名称: | CYS-TYR-ARG-LYS-LEU, Periplasmic serine endoprotease DegP | | 著者 | Cho, H, Choi, Y, Lee, H.H, Kim, S. | | 登録日 | 2019-02-26 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (4.2 Å) | | 主引用文献 | Over-activation of a nonessential bacterial protease DegP as an antibiotic strategy.
Commun Biol, 3, 2020
|
|
6JJK
 
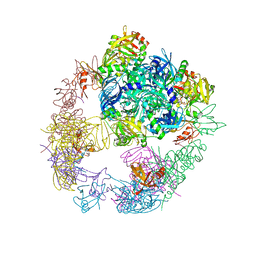 | | Crystal structure of the DegP dodecamer with a modulator | | 分子名称: | CYS-TYR-TYR-LYS-ILE, Periplasmic serine endoprotease DegP | | 著者 | Cho, H, Choi, Y, Lee, H.H, Kim, S. | | 登録日 | 2019-02-26 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Over-activation of a nonessential bacterial protease DegP as an antibiotic strategy
Commun Biol, 3, 2020
|
|
