2YK9
 
 | | Tricyclic series of Hsp90 inhibitors | | Descriptor: | 4-(1H-IMIDAZO[4,5-C]PYRIDIN-2-YL)FLUOREN-9-ONE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Dupuy, A, Vallee, F. | | Deposit date: | 2011-05-26 | | Release date: | 2011-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Tricyclic Series of Heat Shock Protein 90 (Hsp90) Inhibitors Part I: Discovery of Tricyclic Imidazo[4,5-C]Pyridines as Potent Inhibitors of the Hsp90 Molecular Chaperone.
J.Med.Chem., 54, 2011
|
|
2YKC
 
 | | Tricyclic series of Hsp90 inhibitors | | Descriptor: | HEAT SHOCK PROTEIN HSP 90-ALPHA, N-[4-(3H-IMIDAZO[4,5-C]PYRIDIN-2-YL)-9H-FLUOREN-9-YL-ISONICOTINAMIDE | | Authors: | Dupuy, A, Vallee, F. | | Deposit date: | 2011-05-26 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Tricyclic Series of Heat Shock Protein 90 (Hsp90) Inhibitors Part I: Discovery of Tricyclic Imidazo[4,5-C]Pyridines as Potent Inhibitors of the Hsp90 Molecular Chaperone.
J.Med.Chem., 54, 2011
|
|
2YKB
 
 | | Tricyclic series of Hsp90 inhibitors | | Descriptor: | HEAT SHOCK PROTEIN HSP 90-ALPHA, N-[4-(3H-IMIDAZO[4,5-C]PYRIDIN-2-YL)-9H-FLUOREN-9-YL]-SUCCINAMIDE | | Authors: | Dupuy, A, Vallee, F. | | Deposit date: | 2011-05-26 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Tricyclic Series of Heat Shock Protein 90 (Hsp90) Inhibitors Part I: Discovery of Tricyclic Imidazo[4,5-C]Pyridines as Potent Inhibitors of the Hsp90 Molecular Chaperone.
J.Med.Chem., 54, 2011
|
|
2YKE
 
 | | Tricyclic series of Hsp90 inhibitors | | Descriptor: | HEAT SHOCK PROTEIN HSP 90-ALPHA, N-[(4R)-4-(3H-imidazo[4,5-c]pyridin-2-yl)-4H-fluoren-9-yl]quinoline-5-carboxamide | | Authors: | Dupuy, A, Vallee, F. | | Deposit date: | 2011-05-26 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Tricyclic Series of Heat Shock Protein 90 (Hsp90) Inhibitors Part I: Discovery of Tricyclic Imidazo[4,5-C]Pyridines as Potent Inhibitors of the Hsp90 Molecular Chaperone.
J.Med.Chem., 54, 2011
|
|
2YKI
 
 | | Tricyclic series of Hsp90 inhibitors | | Descriptor: | 1-H-PYRROLO[2,3-B]PYRIDINE-4-CARBOXYLIC ACID [4-(3H-IMIDAZO[4,5-C]PYRIDIN-2-YL)-9H-FLUOREN-9-YL]-AMIDE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Dupuy, A, Vallee, F. | | Deposit date: | 2011-05-27 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Tricyclic Series of Heat Shock Protein 90 (Hsp90) Inhibitors Part I: Discovery of Tricyclic Imidazo[4,5-C]Pyridines as Potent Inhibitors of the Hsp90 Molecular Chaperone.
J.Med.Chem., 54, 2011
|
|
2Q5Q
 
 | | X-ray structure of phenylpyruvate decarboxylase in complex with 3-deaza-ThDP and 5-phenyl-2-oxo-valeric acid | | Descriptor: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-3-METHYLTHIOPHEN-2-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, 5-PHENYL-2-KETO-VALERIC ACID, GLYCEROL, ... | | Authors: | Versees, W, Spaepen, S, Wood, M.D, Leeper, F.J, Vanderleyden, J, Steyaert, J. | | Deposit date: | 2007-06-01 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular mechanism of allosteric substrate activation in a thiamine diphosphate-dependent decarboxylase.
J.Biol.Chem., 282, 2007
|
|
2Q5L
 
 | | X-ray structure of phenylpyruvate decarboxylase in complex with 2-(1-hydroxyethyl)-3-deaza-ThDP | | Descriptor: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-5-[(1R)-1-HYDROXYETHYL]-3-METHYL-2-THIENYL}ETHYL TRIHYDROGEN DIPHOSPHATE, 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-5-[(1S)-1-HYDROXYETHYL]-3-METHYL-2-THIENYL}ETHYL TRIHYDROGEN DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Versees, W, Spaepen, S, Wood, M.D, Leeper, F.J, Vanderleyden, J, Steyaert, J. | | Deposit date: | 2007-06-01 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular mechanism of allosteric substrate activation in a thiamine diphosphate-dependent decarboxylase.
J.Biol.Chem., 282, 2007
|
|
2Q5J
 
 | | X-ray structure of phenylpyruvate decarboxylase in complex with 3-deaza-ThDP | | Descriptor: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-3-METHYLTHIOPHEN-2-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, MAGNESIUM ION, Phenylpyruvate decarboxylase | | Authors: | Versees, W, Spaepen, S, Wood, M.D, Leeper, F.J, Vanderleyden, J, Steyaert, J. | | Deposit date: | 2007-06-01 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular mechanism of allosteric substrate activation in a thiamine diphosphate-dependent decarboxylase.
J.Biol.Chem., 282, 2007
|
|
2Q5O
 
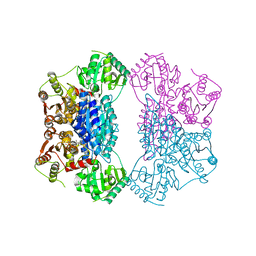 | | X-ray structure of phenylpyruvate decarboxylase in complex with 3-deaza-ThDP and phenylpyruvate | | Descriptor: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-3-METHYLTHIOPHEN-2-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, 3-PHENYLPYRUVIC ACID, GLYCEROL, ... | | Authors: | Versees, W, Spaepen, S, Wood, M.D, Leeper, F.J, Vanderleyden, J, Steyaert, J. | | Deposit date: | 2007-06-01 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular mechanism of allosteric substrate activation in a thiamine diphosphate-dependent decarboxylase.
J.Biol.Chem., 282, 2007
|
|
2VBF
 
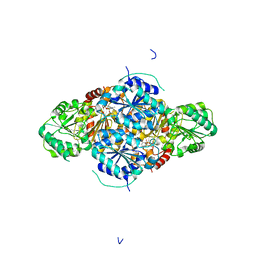 | | The holostructure of the branched-chain keto acid decarboxylase (KdcA) from Lactococcus lactis | | Descriptor: | BRANCHED-CHAIN ALPHA-KETOACID DECARBOXYLASE, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Berthold, C.L, Gocke, D, Wood, M.D, Leeper, F, Pohl, M, Schneider, G. | | Deposit date: | 2007-09-12 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Branched-Chain Keto Acid Decarboxylase (Kdca) from Lactococcus Lactis Provides Insights Into the Structural Basis for the Chemo- and Enantioselective Carboligation Reaction
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2VBG
 
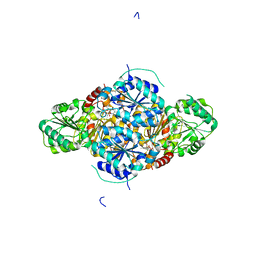 | | The complex structure of the branched-chain keto acid decarboxylase (KdcA) from Lactococcus lactis with 2R-1-hydroxyethyl-deazaThDP | | Descriptor: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-5-[(1R)-1-HYDROXYETHYL]-3-METHYL-2-THIENYL}ETHYL TRIHYDROGEN DIPHOSPHATE, BRANCHED-CHAIN ALPHA-KETOACID DECARBOXYLASE, MAGNESIUM ION | | Authors: | Berthold, C.L, Gocke, D, Wood, M.D, Leeper, F, Pohl, M, Schneider, G. | | Deposit date: | 2007-09-12 | | Release date: | 2007-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Branched-Chain Keto Acid Decarboxylase (Kdca) from Lactococcus Lactis Provides Insights Into the Structural Basis for the Chemo- and Enantioselective Carboligation Reaction
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1HY1
 
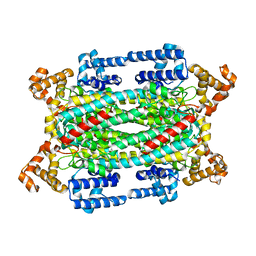 | |
1I0A
 
 | |
1HY0
 
 | | CRYSTAL STRUCTURE OF WILD TYPE DUCK DELTA 1 CRYSTALLIN (EYE LENS PROTEIN) | | Descriptor: | DELTA CRYSTALLIN I, SULFATE ION | | Authors: | Sampaleanu, L.M, Vallee, F, Slingsby, C, Howell, P.L. | | Deposit date: | 2001-01-17 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural studies of duck delta 1 and delta 2 crystallin suggest conformational changes occur during catalysis.
Biochemistry, 40, 2001
|
|
3ZLS
 
 | | Crystal structure of MEK1 in complex with fragment 6 | | Descriptor: | 1H-PYRROLO[2,3-B]PYRIDINE-3-CARBOXYLIC ACID, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1, SODIUM ION | | Authors: | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3ZM4
 
 | | Crystal structure of MEK1 in complex with fragment 1 | | Descriptor: | 7-chloranyl-6-[(3S)-pyrrolidin-3-yl]oxy-2H-isoquinolin-1-one, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1 | | Authors: | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | Deposit date: | 2013-02-05 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3ZLW
 
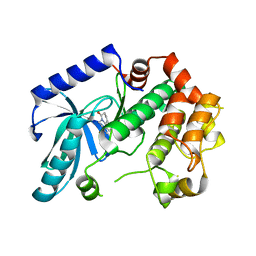 | | Crystal structure of MEK1 in complex with fragment 3 | | Descriptor: | (1R)-1-hydroxy-1-methyl-2,3,6,7-tetrahydro-1H,5H-pyrido[3,2,1-ij]quinolin-5-one, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1 MAPK/ERK KINASE 1, MEK 1, ... | | Authors: | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2JI7
 
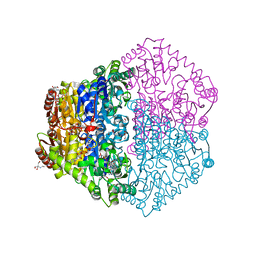 | | X-ray structure of Oxalyl-CoA decarboxylase with covalent reaction intermediate | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-{(1R,11R,15S,17R)-19-[(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]-1,11,15,17-TETRAHYDROXY-12,12-DIMETHYL-15,17-DIOXIDO-6,10-DIOXO-14,16,18-TRIOXA-2-THIA-5,9-DIAZA-15,17-DIPHOSPHANONADEC-1-YL}-5-(2-{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Berthold, C.L, Toyota, C.G, Moussatche, P, Wood, M.D, Leeper, F, Richards, N.G.J, Lindqvist, Y. | | Deposit date: | 2007-02-26 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystallographic Snapshots of Oxalyl-Coa Decarboxylase Give Insights Into Catalysis by Nonoxidative Thdp-Dependent Decarboxylases
Structure, 15, 2007
|
|
2JIB
 
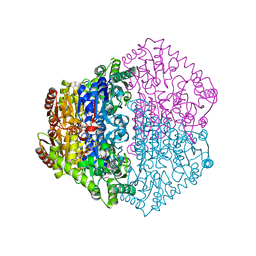 | | X-ray structure of Oxalyl-CoA decarboxylase in complex with coenzyme- A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, COENZYME A, MAGNESIUM ION, ... | | Authors: | Berthold, C.L, Toyota, C.G, Moussatche, P, Wood, M.D, Leeper, F, Richards, N.G.J, Lindqvist, Y. | | Deposit date: | 2007-02-26 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic Snapshots of Oxalyl-Coa Decarboxylase Give Insights Into Catalysis by Nonoxidative Thdp-Dependent Decarboxylases
Structure, 15, 2007
|
|
2JI6
 
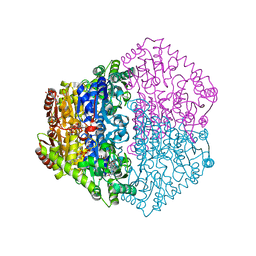 | | X-ray structure of Oxalyl-CoA decarboxylase in complex with 3-deaza- ThDP and oxalyl-CoA | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-3-METHYLTHIOPHEN-2-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Berthold, C.L, Toyota, C.G, Moussatche, P, Wood, M.D, Leeper, F, Richards, N.G.J, Lindqvist, Y. | | Deposit date: | 2007-02-26 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystallographic Snapshots of Oxalyl-Coa Decarboxylase Give Insights Into Catalysis by Nonoxidative Thdp-Dependent Decarboxylases
Structure, 15, 2007
|
|
2JI9
 
 | | X-ray structure of Oxalyl-CoA decarboxylase in complex with 3-deaza- ThDP | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-3-METHYLTHIOPHEN-2-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Berthold, C.L, Toyota, C.G, Moussatche, P, Wood, M.D, Leeper, F, Richards, N.G.J, Lindqvist, Y. | | Deposit date: | 2007-02-26 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic Snapshots of Oxalyl-Coa Decarboxylase Give Insights Into Catalysis by Nonoxidative Thdp-Dependent Decarboxylases
Structure, 15, 2007
|
|
4KXY
 
 | | Human transketolase in complex with ThDP analogue (R)-2-(1,2-dihydroxyethyl)-3-deaza-ThDP | | Descriptor: | 1,2-ETHANEDIOL, 2-{4-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-[(1R)-1,2-dihydroxyethyl]-3-methylthiophen-2-yl}ethyl trihydrogen diphosphate, CALCIUM ION, ... | | Authors: | Neumann, P, Luedtke, S, Erixon, K.M, Leeper, F, Kluger, R, Ficner, R, Tittmann, K. | | Deposit date: | 2013-05-28 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Sub-angstrom-resolution crystallography reveals physical distortions that enhance reactivity of a covalent enzymatic intermediate.
Nat Chem, 5, 2013
|
|
2JI8
 
 | | X-ray structure of Oxalyl-CoA decarboxylase in complex with Formyl- CoA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, OXALYL-COA DECARBOXYLASE, ... | | Authors: | Berthold, C.L, Toyota, C.G, Moussatche, P, Wood, M.D, Leeper, F, Richards, N.G.J, Lindqvist, Y. | | Deposit date: | 2007-02-26 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic Snapshots of Oxalyl-Coa Decarboxylase Give Insights Into Catalysis by Nonoxidative Thdp-Dependent Decarboxylases
Structure, 15, 2007
|
|
3ZLY
 
 | | Crystal structure of MEK1 in complex with fragment 8 | | Descriptor: | 3-AMINO-1H-INDAZOLE-4-CARBONITRILE, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1, SODIUM ION | | Authors: | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3ZLX
 
 | | Crystal structure of MEK1 in complex with fragment 18 | | Descriptor: | 7-choro-6-[(3R)-pyrrolidin-3-ylmethoxy]isoquinolin-1(2H)-one, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1 | | Authors: | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
