1MT4
 
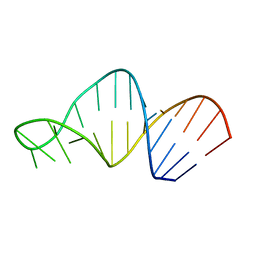 | | Structure of 23S ribosomal RNA hairpin 35 | | Descriptor: | 23S ribosomal Hairpin 35 | | Authors: | Lebars, I, Yoshizawa, S, Stenholm, A.R, Guittet, E, Douthwaite, S, Fourmy, D. | | Deposit date: | 2002-09-20 | | Release date: | 2003-01-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of 23S rRNA hairpin 35 and its interaction with the tylosin-resistance methyltransferase RlmAII
Embo J., 22, 2003
|
|
2OOM
 
 | | NMR structure of a kissing complex formed between the TAR RNA element of HIV-1 and a LNA/RNA aptamer | | Descriptor: | RNA 16-mer, TAR RNA element of HIV-1 | | Authors: | Lebars, I, Richard, T, Di Primo, C, Toulme, J.J. | | Deposit date: | 2007-01-26 | | Release date: | 2008-01-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a kissing complex formed between the TAR RNA element of HIV-1 and a LNA-modified aptamer
Nucleic Acids Res., 35, 2007
|
|
2PN9
 
 | | NMR structure of a kissing complex formed between the TAR RNA element of HIV-1 and a LNA modified aptamer | | Descriptor: | 5'-R(*GP*GP*AP*GP*CP*CP*UP*GP*GP*GP*AP*GP*CP*UP*CP*C)-3', RNA 16-mer with locked residues 9-10 | | Authors: | Lebars, I, Richard, T, Di Primo, C, Toulme, J.-J. | | Deposit date: | 2007-04-24 | | Release date: | 2007-10-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a kissing complex formed between the TAR RNA element of HIV-1 and a LNA-modified aptamer
Nucleic Acids Res., 35, 2007
|
|
1K6H
 
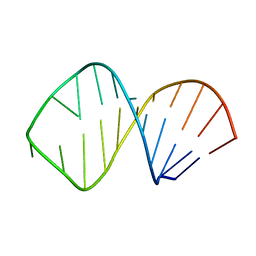 | | Solution Structure of Conserved AGNN Tetraloops: Insights into Rnt1p RNA processing | | Descriptor: | RNA (5'-R(P*GP*GP*CP*GP*UP*GP*UP*UP*CP*AP*GP*AP*AP*GP*AP*AP*CP*GP*CP*GP*CP*C)-3') | | Authors: | Lebars, I, Lamontagne, B, Yoshizawa, S, Abou Elela, S, Fourmy, D. | | Deposit date: | 2001-10-16 | | Release date: | 2001-12-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of conserved AGNN tetraloops: insights into Rnt1p RNA processing.
EMBO J., 20, 2001
|
|
1K6G
 
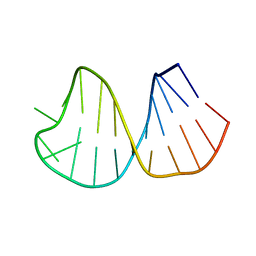 | | Solution Structure of Conserved AGNN Tetraloops: Insights into Rnt1p RNA Processing | | Descriptor: | RNA (5'-R(P*GP*GP*CP*GP*UP*CP*AP*UP*GP*AP*GP*UP*CP*CP*AP*UP*GP*GP*CP*GP*CP*C)-3') | | Authors: | Lebars, I, Lamontagne, B, Yoshizawa, S, Abou Elela, S, Fourmy, D. | | Deposit date: | 2001-10-16 | | Release date: | 2001-12-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of conserved AGNN tetraloops: insights into Rnt1p RNA processing.
EMBO J., 20, 2001
|
|
6HYK
 
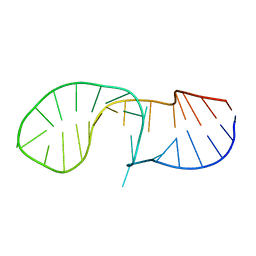 | | NMR solution structure of the C/D box snoRNA U14 | | Descriptor: | RNA (31-MER) | | Authors: | Chagot, M.E, Quinternet, M, Rothe, B, Charpentier, B, Coutant, J, Manival, X, Lebars, I. | | Deposit date: | 2018-10-22 | | Release date: | 2019-04-24 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The yeast C/D box snoRNA U14 adopts a "weak" K-turn like conformation recognized by the Snu13 core protein in solution.
Biochimie, 164, 2019
|
|
5IEM
 
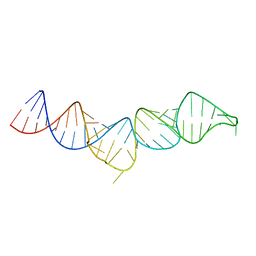 | | NMR structure of the 5'-terminal hairpin of the 7SK snRNA | | Descriptor: | 7SK snRNA | | Authors: | Bourbigot, S, Dock-Bregeon, A.C, Coutant, J, Kieffer, B, Lebars, I. | | Deposit date: | 2016-02-25 | | Release date: | 2016-11-02 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 5'-terminal hairpin of the 7SK small nuclear RNA.
RNA, 22, 2016
|
|
1GYZ
 
 | |
