8SGB
 
 | |
4GUP
 
 | | Structure of MHC-class I related molecule MR1 | | Descriptor: | 2-amino-4-oxo-3,4-dihydropteridine-6-carbaldehyde, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Patel, O, Le Nours, J, Rossjohn, J. | | Deposit date: | 2012-08-29 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | MR1 presents microbial vitamin B metabolites to MAIT cells
Nature, 491, 2012
|
|
2WZT
 
 | | Crystal structure of a mycobacterium aldo-keto reductase in its apo and liganded form | | Descriptor: | ALDO-KETO REDUCTASE | | Authors: | Scoble, J, McAlister, A.D, Fulton, Z, Troy, S, Byres, E, Vivian, J.P, Brammananth, R, Wilce, M.C.J, Le Nours, J, Zaker-Tabrizi, L, Coppel, R.L, Crellin, P.K, Rossjohn, J, Beddoe, T. | | Deposit date: | 2009-12-03 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure and Comparative Functional Analyses of a Mycobacterium Aldo-Keto Reductase.
J.Mol.Biol., 398, 2010
|
|
7RYN
 
 | | CD1a-sulfatide-gdTCR complex | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wegrecki, M, Le Nours, J, Rossjohn, J. | | Deposit date: | 2021-08-25 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Atypical sideways recognition of CD1a by autoreactive gamma delta T cell receptors.
Nat Commun, 13, 2022
|
|
7RYL
 
 | | T cell receptor CO3 | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, T cell receptor delta variable 1,T cell receptor alpha chain constant, ... | | Authors: | Wegrecki, M, Le Nours, J, Rossjohn, J. | | Deposit date: | 2021-08-25 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atypical sideways recognition of CD1a by autoreactive gamma delta T cell receptors.
Nat Commun, 13, 2022
|
|
7RYM
 
 | | CD1a-endo-gdTCR complex | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Wegrecki, M, Le Nours, J, Rossjohn, J. | | Deposit date: | 2021-08-25 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Atypical sideways recognition of CD1a by autoreactive gamma delta T cell receptors.
Nat Commun, 13, 2022
|
|
7RYO
 
 | | CD1a-dideoxymycobactin-gdTCR complex | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, SULFATE ION, ... | | Authors: | Wegrecki, M, Le Nours, J, Rossjohn, J. | | Deposit date: | 2021-08-25 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Atypical sideways recognition of CD1a by autoreactive gamma delta T cell receptors.
Nat Commun, 13, 2022
|
|
4APQ
 
 | | Crystal structure of autoreactive-Valpha14-Vbeta6 NKT TCR in complex with CD1d-sulfatide | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIGEN-PRESENTING GLYCOPROTEIN CD1D1, ... | | Authors: | Clarke, A.J, Le Nours, J, Rossjohn, J. | | Deposit date: | 2012-04-05 | | Release date: | 2013-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Type-II Natural Killer T Cell Antigen Receptor Mediated Recognition of Cd1D-Sulfatide
To be Published
|
|
4WNQ
 
 | | THE MOLECULAR BASES OF DELTA/ALPHA-BETA T-CELL MEDIATED ANTIGEN RECOGNITION | | Descriptor: | TCR Variable Beta 2 (TRBV20) chain and TCR constant Beta chain, TCR Variable Delta 1 chain and TCR constant Alpha chain | | Authors: | Pellicci, D.G, Uldrich, A.P, Le Nours, J, Ross, F, Chabrol, E, Eckle, S.B.G, de Boer, R, Lim, R.T, McPherson, K, Besra, G, Howell, A.R, Moretta, L, McCluskey, J, Heemskerk, M.H.M, Gras, S, Rossjohn, J, Godfrey, D.I. | | Deposit date: | 2014-10-14 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular bases of delta / alpha beta T cell-mediated antigen recognition.
J.Exp.Med., 211, 2014
|
|
4WO4
 
 | | The molecular bases of Delta/Alpha beta T cell-mediated antigen recognition. | | Descriptor: | (15Z)-N-[(2S,3S,4R)-1-(alpha-D-galactopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]tetracos-15-enamide, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pellicci, D.G, Uldrich, A.P, Le Nours, J, Ross, F, Chabrol, E, Eckle, S.B.G, de Boer, R, Lim, R.T, McPherson, K, Besra, G, Howell, A.R, Moretta, L, McCluskey, J, Heemskerk, M.H.M, Gras, S, Rossjohn, J, Godfrey, D.I. | | Deposit date: | 2014-10-15 | | Release date: | 2014-11-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The molecular bases of delta / alpha beta T cell-mediated antigen recognition.
J.Exp.Med., 211, 2014
|
|
6MSS
 
 | | Diversity in the type II Natural Killer T cell receptor repertoire and antigen specificity leads to differing CD1d docking strategies | | Descriptor: | (2S)-2-(heptadecanoyloxy)-3-{[(10S)-10-methyloctadecanoyl]oxy}propyl alpha-D-glucopyranosiduronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sundararaj, S, Le Nours, J, Praveena, T, Rossjohn, J. | | Deposit date: | 2018-10-18 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Distinct CD1d docking strategies exhibited by diverse Type II NKT cell receptors.
Nat Commun, 10, 2019
|
|
6MRA
 
 | | Diversity in the type II Natural Killer T cell receptor repertoire and antigen specificity leads to differing CD1d docking strategies | | Descriptor: | TCR alpha chain, TCR beta-chain | | Authors: | Sundararaj, S, Le Nours, J, Praveena, T, Rossjohn, J. | | Deposit date: | 2018-10-12 | | Release date: | 2019-09-18 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Distinct CD1d docking strategies exhibited by diverse Type II NKT cell receptors.
Nat Commun, 10, 2019
|
|
6NUX
 
 | | CD1a-lipid binary complex | | Descriptor: | (2E,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, 1,2-ETHANEDIOL, Beta-2-microglobulin, ... | | Authors: | Wegrecki, M, Le Nours, J, Rossjohn, J. | | Deposit date: | 2019-02-03 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human T cell response to CD1a and contact dermatitis allergens in botanical extracts and commercial skin care products.
Sci Immunol, 5, 2020
|
|
6BMH
 
 | | Crystal structure of MHC-I like protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d2, ... | | Authors: | Khandokar, Y.B, Le Nours, J, Rossjohn, J. | | Deposit date: | 2017-11-14 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Differing roles of CD1d2 and CD1d1 proteins in type I natural killer T cell development and function.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BMK
 
 | | Crystal structure of MHC-I like protein | | Descriptor: | (2R)-1-(decanoyloxy)-3-(phosphonooxy)propan-2-yl octadecanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Khandokar, Y.B, Le Nours, J, Rossjohn, J. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Differing roles of CD1d2 and CD1d1 proteins in type I natural killer T cell development and function.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7K0X
 
 | | Marsupial T cell receptor Spl_145 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, T cell receptor gamma chain, T cell receptor mu chain, ... | | Authors: | Wegrecki, M, Kannan Sivaraman, K, Le Nours, J, Rossjohn, J. | | Deposit date: | 2020-09-06 | | Release date: | 2021-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The molecular assembly of the marsupial gamma mu T cell receptor defines a third T cell lineage.
Science, 371, 2021
|
|
1R8L
 
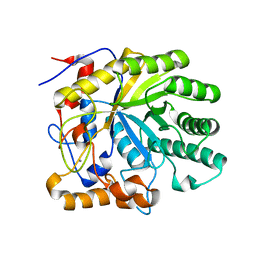 | | The structure of endo-beta-1,4-galactanase from Bacillus licheniformis | | Descriptor: | CALCIUM ION, endo-beta-1,4-galactanase | | Authors: | Ryttersgaard, C, Le Nours, J, Lo Leggio, L, Jorgensen, C.T, Christensen, L.L, Bjornvad, M, Larsen, S. | | Deposit date: | 2003-10-27 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products
J.Mol.Biol., 341, 2004
|
|
7KP1
 
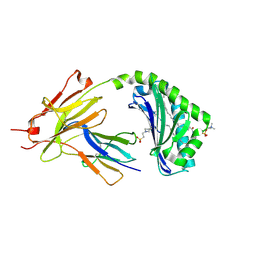 | | CD1a-42:2 SM binary complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-2-microglobulin, T-cell surface glycoprotein CD1a, ... | | Authors: | Wegrecki, M, Le Nours, J, Rossjohn, J. | | Deposit date: | 2020-11-10 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | CD1a selectively captures endogenous cellular lipids that broadly block T cell response.
J.Exp.Med., 218, 2021
|
|
7KP0
 
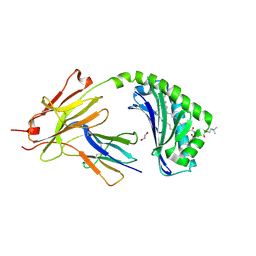 | | CD1a-42:1 SM binary complex | | Descriptor: | (4R,7S)-4-hydroxy-7-[(1S,2E)-1-hydroxyhexadec-2-en-1-yl]-N,N,N-trimethyl-4,9-dioxo-3,5-dioxa-8-aza-4lambda~5~-phosphadotriacontan-1-aminium, 1,2-ETHANEDIOL, Beta-2-microglobulin, ... | | Authors: | Wegrecki, M, Le Nours, J, Rossjohn, J. | | Deposit date: | 2020-11-10 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CD1a selectively captures endogenous cellular lipids that broadly block T cell response.
J.Exp.Med., 218, 2021
|
|
7KOZ
 
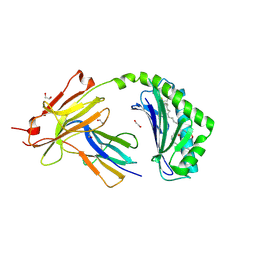 | | CD1a-36:2 SM binary complex | | Descriptor: | (4S,7S,17Z)-4-hydroxy-7-[(1S,2E)-1-hydroxyhexadec-2-en-1-yl]-N,N,N-trimethyl-4,9-dioxo-3,5-dioxa-8-aza-4lambda~5~-phosphahexacos-17-en-1-aminium, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wegrecki, M, Le Nours, J, Rossjohn, J. | | Deposit date: | 2020-11-10 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | CD1a selectively captures endogenous cellular lipids that broadly block T cell response.
J.Exp.Med., 218, 2021
|
|
1UR4
 
 | | The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products. | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GALACTANASE, ... | | Authors: | Ryttersgaard, C, Le Nours, J, Lo Leggio, L, Jorgensen, C.T, Christensen, L.L.H, Bjornvad, M, Larsen, S. | | Deposit date: | 2003-10-24 | | Release date: | 2004-10-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of Endo-Beta-1,4-Galactanase from Bacillus Licheniformis in Complex with Two Oligosaccharide Products
J.Mol.Biol., 341, 2004
|
|
1UR0
 
 | | The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products. | | Descriptor: | CALCIUM ION, GALACTANASE, TRIETHYLENE GLYCOL, ... | | Authors: | Ryttersgaard, C, Le Nours, J, Lo Leggio, L, Jorgensen, C.T, Christensen, L.L.H, Bjornvad, M, Larsen, S. | | Deposit date: | 2003-10-24 | | Release date: | 2004-10-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structure of Endo-Beta-1,4-Galactanase from Bacillus Licheniformis in Complex with Two Oligosaccharide Products
J.Mol.Biol., 341, 2004
|
|
7K0Z
 
 | | Marsupial T cell receptor Spl_157 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, T cell receptor gamma chain, T cell receptor mu chain | | Authors: | Wegrecki, M, Kannan Sivaraman, K, Le Nours, J, Rossjohn, J. | | Deposit date: | 2020-09-06 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The molecular assembly of the marsupial gamma mu T cell receptor defines a third T cell lineage.
Science, 371, 2021
|
|
7L15
 
 | | Marsupial T cell receptor Spl_118 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, T cell receptor gamma chain, ... | | Authors: | Wegrecki, M, Kannan Sivaraman, K, Praveena, T, Le Nours, J, Rossjohn, J. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The molecular assembly of the marsupial gamma mu T cell receptor defines a third T cell lineage.
Science, 371, 2021
|
|
1UUM
 
 | | Rat dihydroorotate dehydrogenase (DHOD)in complex with atovaquone | | Descriptor: | 2-[4-(4-CHLOROPHENYL)CYCLOHEXYLIDENE]-3,4-DIHYDROXY-1(2H)-NAPHTHALENONE, DIHYDROOROTATE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Hansen, M, Le Nours, J, Johansson, E, Antal, T, Ullrich, A, Loffler, M, Larsen, S. | | Deposit date: | 2004-01-06 | | Release date: | 2004-04-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibitor Binding in a Class 2 Dihydroorotate Dehydrogenase Causes Variations in the Membrane-Associated N-Terminal Domain
Protein Sci., 13, 2004
|
|
