6ZUQ
 
 | | Crystal structure of the effector Ecp11-1 from Fulvia fulva | | Descriptor: | Extracellular protein 11-1, GLYCEROL, ZINC ION | | Authors: | Lazar, N, Mesarich, C, Petit-Houdenot, Y, Talbi, N, Li de la Sierra-Gallay, I, Zelie, E, Blondeau, K, Gracy, J, Ollivier, B, van de Wouw, A, Balesdent, M.H, Idnurm, A, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2020-07-23 | | Release date: | 2021-08-04 | | Last modified: | 2022-07-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins.
Plos Pathog., 18, 2022
|
|
6ZUS
 
 | | Crystal structure of the effector Ecp11-1 from Fulvia fulva | | Descriptor: | DI(HYDROXYETHYL)ETHER, Extracellular protein 11-1, GLYCEROL, ... | | Authors: | Lazar, N, Mesarich, C, Petit-Houdenot, Y, Talbi, N, Li de la Sierra-Gallay, I, Zelie, E, Blondeau, K, Gracy, J, Ollivier, B, van de Wouw, A, Balesdent, M.H, Idnurm, A, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2020-07-23 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins.
Plos Pathog., 18, 2022
|
|
7AD5
 
 | | Crystal structure of the effector AvrLm5-9 from Leptosphaeria maculans | | Descriptor: | ACETATE ION, Avirulence protein LmJ1, GLYCEROL, ... | | Authors: | Lazar, N, Mesarich, C, Petit-Houdenot, Y, Talbi, N, Li de la Sierra-Gallay, I, Zelie, E, Blondeau, K, Gracy, J, Ollivier, B, van de Wouw, A, Balesdent, M.H, Idnurm, A, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2020-09-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins.
Plos Pathog., 18, 2022
|
|
7B76
 
 | | Crystal structure of the effector AvrLm5-9 from Leptosphaeria maculans | | Descriptor: | Avirulence protein LmJ1, IODIDE ION | | Authors: | Lazar, N, Mesarich, C, Petit-Houdenot, Y, Talbi, N, Li de la Sierra-Gallay, I, Zelie, E, Blondeau, K, Gracy, J, Ollivier, B, van de Wouw, A, Balesdent, M.H, Idnurm, A, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2020-12-09 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins.
Plos Pathog., 18, 2022
|
|
5CM2
 
 | | Structure of Y. lipolytica Trm9-Trm112 complex, a methyltransferase modifying U34 in the anticodon loop of some tRNAs | | Descriptor: | TRNA METHYLTRANSFERASE, TRNA METHYLTRANSFERASE ACTIVATOR SUBUNIT, ZINC ION | | Authors: | Letoquart, J, van Tran, N, Caroline, V, Aleksandrov, A, Lazar, N, Van Tilbeurgh, H, Liger, D, Graille, M. | | Deposit date: | 2015-07-16 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into molecular plasticity in protein complexes from Trm9-Trm112 tRNA modifying enzyme crystal structure.
Nucleic Acids Res., 43, 2015
|
|
4AKK
 
 | | Structure of the NasR transcription antiterminator | | Descriptor: | 1,2-ETHANEDIOL, NITRATE REGULATORY PROTEIN | | Authors: | Boudes, M, Lazar, N, Graille, M, Durand, D, Gaidenko, T.A, Stewart, V, van Tilbeurgh, H. | | Deposit date: | 2012-02-24 | | Release date: | 2012-06-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.145 Å) | | Cite: | The Structure of the Nasr Transcription Antiterminator Reveals a One-Component System with a Nit Nitrate Receptor Coupled to an Antar RNA-Binding Effector.
Mol.Microbiol., 85, 2012
|
|
4A8E
 
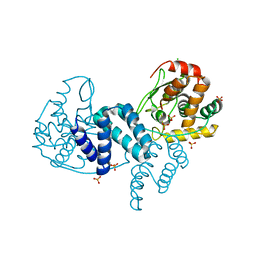 | | The structure of a dimeric Xer recombinase from archaea | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PROBABLE TYROSINE RECOMBINASE XERC-LIKE, ... | | Authors: | Brooks, M.A, ElArnaout, T, Duranda, D, Lisboa, J, Lazar, N, Raynal, B, vanTilbeurgh, H, Serre, M, Quevillon-Cheruel, S. | | Deposit date: | 2011-11-21 | | Release date: | 2012-12-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The Carboxy-Terminal Alpha N Helix of the Archaeal Xera Tyrosine Recombinase is a Molecular Switch to Control Site-Specific Recombination.
Plos One, 8, 2013
|
|
4PUF
 
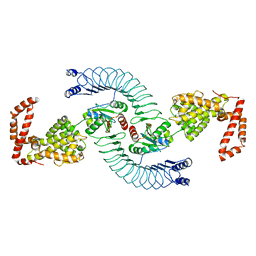 | | Complex between the Salmonella T3SS effector SlrP and its human target thioredoxin-1 | | Descriptor: | E3 ubiquitin-protein ligase SlrP, Thioredoxin | | Authors: | Zouhir, S, Bernal-Bayard, J, Cordero-Alba, M, Cardenal-Munoz, E, Guimaraes, B, Lazar, N, Ramos-Morales, F, Nessler, S. | | Deposit date: | 2014-03-13 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.296 Å) | | Cite: | The structure of the Slrp-Trx1 complex sheds light on the autoinhibition mechanism of the type III secretion system effectors of the NEL family.
Biochem.J., 464, 2014
|
|
4P78
 
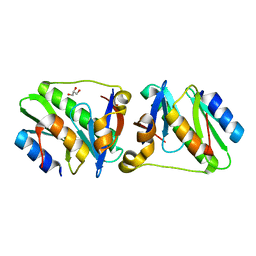 | | HicA3 and HicB3 toxin-antitoxin complex | | Descriptor: | GLYCEROL, HicA3 Toxin, HicB3 antitoxin | | Authors: | Li de la Sierra-Gallay, I, Bibi-Triki, S, van Tilbeurgh, H, Lazar, N, Pradel, E. | | Deposit date: | 2014-03-26 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Functional and Structural Analysis of HicA3-HicB3, a Novel Toxin-Antitoxin System of Yersinia pestis.
J.Bacteriol., 196, 2014
|
|
4P7D
 
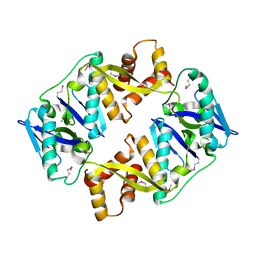 | | Antitoxin HicB3 crystal structure | | Descriptor: | Antitoxin HicB3, CHLORIDE ION | | Authors: | Li de la Sierra-Gallay, I, Bibi-Triki, S, van Tilbeurgh, H, Lazar, N, Pradel, E. | | Deposit date: | 2014-03-27 | | Release date: | 2014-08-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.781 Å) | | Cite: | Functional and Structural Analysis of HicA3-HicB3, a Novel Toxin-Antitoxin System of Yersinia pestis.
J.Bacteriol., 196, 2014
|
|
4OJJ
 
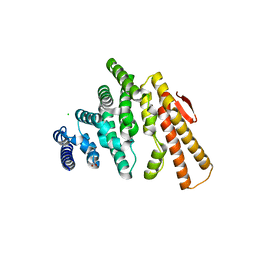 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator (Space group : P212121) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA topoisomerase 2-associated protein PAT1, ... | | Authors: | Fourati-Kammoun, Z, Kolesnikova, O, Back, R, Keller, J, Lazar, N, Gaudon-Plesse, C, Seraphin, B, Graille, M. | | Deposit date: | 2014-01-21 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The C-terminal domain from S. cerevisiae Pat1 displays two conserved regions involved in decapping factor recruitment.
Plos One, 9, 2014
|
|
3P27
 
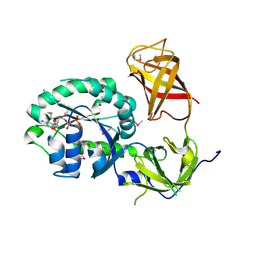 | | Crystal structure of S. cerevisiae Hbs1 protein (GDP-bound form), a translational GTPase involved in RNA quality control pathways and interacting with Dom34/Pelota | | Descriptor: | Elongation factor 1 alpha-like protein, GUANOSINE-5'-DIPHOSPHATE | | Authors: | van den Elzen, A, Henri, J, Lazar, N, Gas, M.E, Durand, D, Lacroute, F, Nicaise, M, van Tilbeurgh, H, Sraphin, B, Graille, M, Paris-Sud Yeast Structural Genomics (YSG) | | Deposit date: | 2010-10-01 | | Release date: | 2010-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Dissection of Dom34-Hbs1 reveals independent functions in two RNA quality control pathways.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4OGP
 
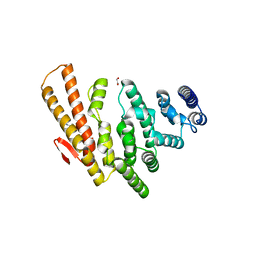 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator (Space group : P21) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA topoisomerase 2-associated protein PAT1 | | Authors: | Fourati-Kammoun, Z, Kolesnikova, O, Back, R, Keller, J, Lazar, N, Gaudon-Plesse, C, Seraphin, B, Graille, M. | | Deposit date: | 2014-01-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The C-terminal domain from S. cerevisiae Pat1 displays two conserved regions involved in decapping factor recruitment.
Plos One, 9, 2014
|
|
3P26
 
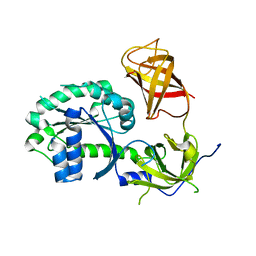 | | Crystal structure of S. cerevisiae Hbs1 protein (apo-form), a translational GTPase involved in RNA quality control pathways and interacting with Dom34/Pelota | | Descriptor: | Elongation factor 1 alpha-like protein | | Authors: | van den Elzen, A, Henri, J, Lazar, N, Gas, M.E, Durand, D, Lacroute, F, Nicaise, M, van Tilbeurgh, H, Sraphin, B, Graille, M, Paris-Sud Yeast Structural Genomics (YSG) | | Deposit date: | 2010-10-01 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dissection of Dom34-Hbs1 reveals independent functions in two RNA quality control pathways.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3Q87
 
 | | Structure of E. cuniculi Mtq2-Trm112 complex responible for the methylation of eRF1 translation termination factor | | Descriptor: | N6 adenine specific DNA methylase, Putative uncharacterized protein ECU08_1170, S-ADENOSYLMETHIONINE, ... | | Authors: | Liger, D, Mora, L, Lazar, N, Figaro, S, Henri, J, Scrima, N, Buckingham, R.H, van Tilbeurgh, H, Heurgue-Hamard, V, Graille, M. | | Deposit date: | 2011-01-06 | | Release date: | 2011-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Mechanism of activation of methyltransferases involved in translation by the Trm112 'hub' protein
Nucleic Acids Res., 2011
|
|
