1THT
 
 | | STRUCTURE OF A MYRISTOYL-ACP-SPECIFIC THIOESTERASE FROM VIBRIO HARVEYI | | Descriptor: | THIOESTERASE | | Authors: | Lawson, D.M, Derewenda, U, Serre, L, Ferri, S, Szitter, R, Wei, Y, Meighen, E.A, Derewenda, Z.S. | | Deposit date: | 1994-04-19 | | Release date: | 1995-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a myristoyl-ACP-specific thioesterase from Vibrio harveyi.
Biochemistry, 33, 1994
|
|
1ATG
 
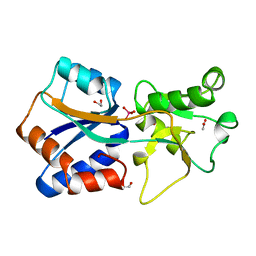 | | AZOTOBACTER VINELANDII PERIPLASMIC MOLYBDATE-BINDING PROTEIN | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, PERIPLASMIC MOLYBDATE-BINDING PROTEIN, ... | | Authors: | Lawson, D.M, Pau, R.N, Williams, C.E.M, Mitchenall, L.A. | | Deposit date: | 1997-08-14 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Ligand size is a major determinant of specificity in periplasmic oxyanion-binding proteins: the 1.2 A resolution crystal structure of Azotobacter vinelandii ModA.
Structure, 6, 1998
|
|
1E84
 
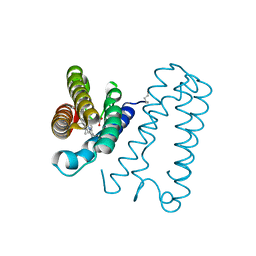 | | Cytochrome c' from Alcaligenes xylosoxidans - reduced structure | | Descriptor: | CYTOCHROME C', HEME C | | Authors: | Lawson, D.M, Stevenson, C.E.M, Andrew, C.R, Eady, R.R. | | Deposit date: | 2000-09-15 | | Release date: | 2000-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unprecedented Proximal Binding of Nitric Oxide to Heme: Implications for Guanylate Cyclase
Embo J., 19, 2000
|
|
1E86
 
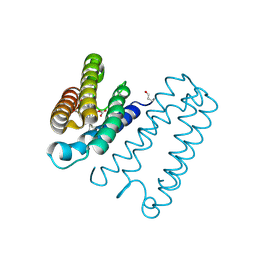 | | Cytochrome c' from Alcaligenes xylosoxidans - reduced structure with CO bound to distal side of heme | | Descriptor: | CARBON MONOXIDE, CYTOCHROME C', HEME C | | Authors: | Lawson, D.M, Stevenson, C.E.M, Andrew, C.R, Eady, R.R. | | Deposit date: | 2000-09-15 | | Release date: | 2000-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unprecedented Proximal Binding of Nitric Oxide to Heme: Implications for Guanylate Cyclase
Embo J., 19, 2000
|
|
1E83
 
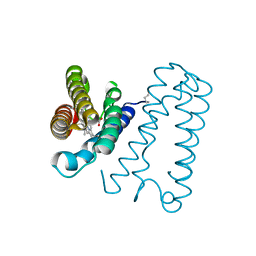 | | Cytochrome c' from Alcaligenes xylosoxidans - oxidized structure | | Descriptor: | CYTOCHROME C', HEME C | | Authors: | Lawson, D.M, Stevenson, C.E.M, Andrew, C.R, Eady, R.R. | | Deposit date: | 2000-09-15 | | Release date: | 2000-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Unprecedented Proximal Binding of Nitric Oxide to Heme: Implications for Guanylate Cyclase
Embo J., 19, 2000
|
|
1E85
 
 | | Cytochrome c' from Alcaligenes xylosoxidans - reduced structure with NO bound to proximal side of heme | | Descriptor: | CYTOCHROME C', HEME C, NITRIC OXIDE | | Authors: | Lawson, D.M, Stevenson, C.E.M, Andrew, C.R, Eady, R.R. | | Deposit date: | 2000-09-15 | | Release date: | 2000-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Unprecedented Proximal Binding of Nitric Oxide to Heme: Implications for Guanylate Cyclase.
Embo J., 19, 2000
|
|
7ANM
 
 | | Nudaurelia capensis omega virus capsid: virus-like particles expressed in Nicotiana benthamiana | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Ribeiro, J.R.S, Domitrovic, T, Hesketh, E.L, Scarff, C.A, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2020-10-12 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Plant-expressed virus-like particles reveal the intricate maturation process of a eukaryotic virus.
Commun Biol, 4, 2021
|
|
7ATA
 
 | | Nudaurelia capensis omega virus procapsid: virus-like particles expressed in Nicotiana benthamiana | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Ribeiro, J.R.S, Domitrovic, T, Hesketh, E.L, Scarff, C.A, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2020-10-29 | | Release date: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (6.63 Å) | | Cite: | Plant-expressed virus-like particles reveal the intricate maturation process of a eukaryotic virus.
Commun Biol, 4, 2021
|
|
8A41
 
 | | Nudaurelia capensis omega virus procapsid at pH7.6 (insect cell expressed VLPs) | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-06-10 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.88 Å) | | Cite: | Nudaurelia capensis omega virus maturation intermediate captured at pH5.9 (insect cell expressed VLPs)
To be published
|
|
8A3C
 
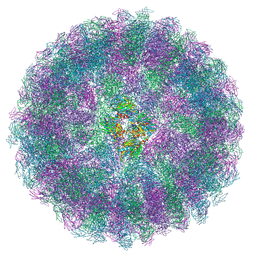 | | Nudaurelia capensis omega virus maturation intermediate captured at pH5.9 (insect cell expressed VLPs) | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-06-08 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Nudaurelia capensis omega virus maturation intermediate captured at pH5.9 (insect cell expressed VLPs)
To be published
|
|
8A6J
 
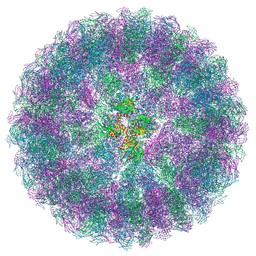 | | Nudaurelia capensis omega virus maturation intermediate captured at pH6.25 (insect cell expressed VLPs) | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-06-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Decoding virus maturation with cryo-EM structures of intermediates
To be published
|
|
3ZT7
 
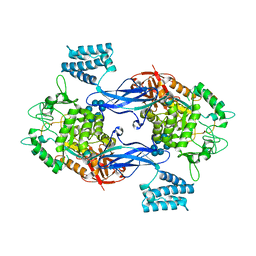 | | GlgE isoform 1 from Streptomyces coelicolor with beta-cyclodextrin and maltose bound | | Descriptor: | Cycloheptakis-(1-4)-(alpha-D-glucopyranose), PUTATIVE GLUCANOHYDROLASE PEP1A, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Syson, K, Stevenson, C.E.M, Rejzek, M, Fairhurst, S.A, Nair, A, Bruton, C.J, Field, R.A, Chater, K.F, Lawson, D.M, Bornemann, S. | | Deposit date: | 2011-07-01 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a Streptomyces Maltosyltransferase Glge: A Homologue of a Genetically Validated Anti-Tuberculosis Target.
J.Biol.Chem., 286, 2011
|
|
3ZT5
 
 | | GlgE isoform 1 from Streptomyces coelicolor with maltose bound | | Descriptor: | PUTATIVE GLUCANOHYDROLASE PEP1A, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Syson, K, Stevenson, C.E.M, Rejzek, M, Fairhurst, S.A, Nair, A, Bruton, C.J, Field, R.A, Chater, K.F, Lawson, D.M, Bornemann, S. | | Deposit date: | 2011-07-01 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of a Streptomyces Maltosyltransferase Glge: A Homologue of a Genetically Validated Anti-Tuberculosis Target.
J.Biol.Chem., 286, 2011
|
|
3ZT6
 
 | | GlgE isoform 1 from Streptomyces coelicolor with alpha-cyclodextrin and maltose bound | | Descriptor: | Cyclohexakis-(1-4)-(alpha-D-glucopyranose), PUTATIVE GLUCANOHYDROLASE PEP1A, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Syson, K, Stevenson, C.E.M, Rejzek, M, Fairhurst, S.A, Nair, A, Bruton, C.J, Field, R.A, Chater, K.F, Lawson, D.M, Bornemann, S. | | Deposit date: | 2011-07-01 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure of a Streptomyces Maltosyltransferase Glge: A Homologue of a Genetically Validated Anti-Tuberculosis Target.
J.Biol.Chem., 286, 2011
|
|
3ZST
 
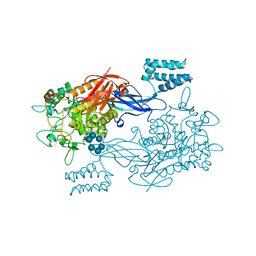 | | GlgE isoform 1 from Streptomyces coelicolor with alpha-cyclodextrin bound | | Descriptor: | 1,2-ETHANEDIOL, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), PUTATIVE GLUCANOHYDROLASE PEP1A GLGE ISOFORM 1 | | Authors: | Syson, K, Stevenson, C.E.M, Rejzek, M, Fairhurst, S.A, Nair, A, Bruton, C.J, Field, R.A, Chater, K.F, Lawson, D.M, Bornemann, S. | | Deposit date: | 2011-06-30 | | Release date: | 2011-09-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a Streptomyces Maltosyltransferase Glge: A Homologue of a Genetically Validated Anti-Tuberculosis Target.
J.Biol.Chem., 286, 2011
|
|
8PFD
 
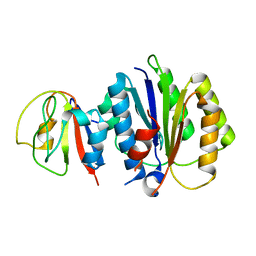 | | Crystal structure of binary complex between Aster yellows witches'-broom phytoplasma effector SAP05 and the von Willebrand Factor Type A domain of the proteasomal ubiquitin receptor Rpn10 from Arabidopsis thaliana | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4 homolog, Sequence-variable mosaic (SVM) signal sequence domain-containing protein | | Authors: | Huang, W, Liu, Q, Maqbool, A, Stevenson, C.E.M, Lawson, D.M, Kamoun, S, Hogenhout, S.A. | | Deposit date: | 2023-06-15 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Bimodular architecture of bacterial effector SAP05 that drives ubiquitin-independent targeted protein degradation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PFC
 
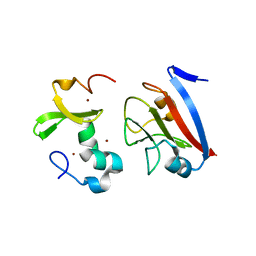 | | Crystal structure of binary complex between Aster yellows witches'-broom phytoplasma effector SAP05 and the zinc finger domain of SPL5 from Arabidopsis thaliana | | Descriptor: | Sequence-variable mosaic (SVM) signal sequence domain-containing protein, Squamosa promoter-binding-like protein 5, ZINC ION | | Authors: | Huang, W, Liu, Q, Maqbool, A, Stevenson, C.E.M, Lawson, D.M, Kamoun, S, Hogenhout, S.A. | | Deposit date: | 2023-06-15 | | Release date: | 2023-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bimodular architecture of bacterial effector SAP05 that drives ubiquitin-independent targeted protein degradation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5CSY
 
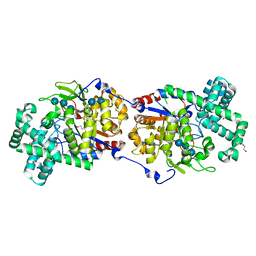 | | Disproportionating enzyme 1 from Arabidopsis - acarbose soak | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase DPE1, chloroplastic/amyloplastic, ... | | Authors: | O'Neill, E.C, Stevenson, C.E.M, Tantanarat, K, Latousakis, D, Donaldson, M.I, Rejzek, M, Limpaseni, T, Smith, A.M, Field, R.A, Lawson, D.M. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1).
J.Biol.Chem., 290, 2015
|
|
5DCW
 
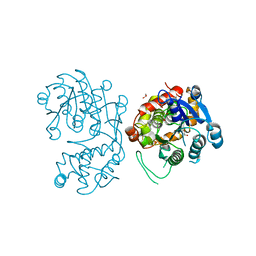 | | Iridoid synthase from Catharanthus roseus - ligand free structure | | Descriptor: | 1,2-ETHANEDIOL, Iridoid synthase | | Authors: | Caputi, L, Kries, H, Stevenson, C.E.M, Kamileen, M.O, Sherden, N.H, Geu-Flores, F, Lawson, D.M, O'Connor, S.E. | | Deposit date: | 2015-08-24 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural determinants of reductive terpene cyclization in iridoid biosynthesis.
Nat.Chem.Biol., 12, 2016
|
|
5CPT
 
 | | Disproportionating enzyme 1 from Arabidopsis - beta cyclodextrin soak | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase DPE1, chloroplastic/amyloplastic, ... | | Authors: | O'Neill, E.C, Stevenson, C.E.M, Tantanarat, K, Latousakis, D, Donaldson, M.I, Rejzek, M, Limpaseni, T, Smith, A.M, Field, R.A, Lawson, D.M. | | Deposit date: | 2015-07-21 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1).
J.Biol.Chem., 290, 2015
|
|
5CVS
 
 | | GlgE isoform 1 from Streptomyces coelicolor E423A mutant soaked in maltoheptaose | | Descriptor: | Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Rashid, A.M, Syson, K, Koliwer-Brandl, H, van de Weerd, R, Stevenson, C.E.M, Batey, S.F.D, Miah, F, Alber, M, Ioerger, T.R, Chandra, G, Appelmelk, B.J, Nartowski, K.P, Khimyak, Y.Z, Lawson, D.M, Jacobs, W.R, Geurtsen, J, Kalscheuer, R, Bornemann, S. | | Deposit date: | 2015-07-27 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ligand-bound structures and site-directed mutagenesis identify the acceptor and secondary binding sites of Streptomyces coelicolor maltosyltransferase GlgE.
J.Biol.Chem., 291, 2016
|
|
5CSU
 
 | | Disproportionating enzyme 1 from Arabidopsis - acarviostatin soak | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase DPE1, chloroplastic/amyloplastic, ... | | Authors: | O'Neill, E.C, Stevenson, C.E.M, Tantanarat, K, Latousakis, D, Donaldson, M.I, Rejzek, M, Limpaseni, T, Smith, A.M, Field, R.A, Lawson, D.M. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1).
J.Biol.Chem., 290, 2015
|
|
5DCU
 
 | | Iridoid synthase from Catharanthus roseus - ternary complex with NADP+ and triethylene glycol carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, Iridoid synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Caputi, L, Kries, H, Stevenson, C.E.M, Kamileen, M.O, Sherden, N.H, Geu-Flores, F, Lawson, D.M, O'Connor, S.E. | | Deposit date: | 2015-08-24 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural determinants of reductive terpene cyclization in iridoid biosynthesis.
Nat.Chem.Biol., 12, 2016
|
|
5DF1
 
 | | Iridoid synthase from Catharanthus roseus - ternary complex with NADP+ and geranic acid | | Descriptor: | (2E)-3,7-dimethylocta-2,6-dienoic acid, 1,2-ETHANEDIOL, IMIDAZOLE, ... | | Authors: | Caputi, L, Kries, H, Stevenson, C.E.M, Kamileen, M.O, Sherden, N.H, Geu-Flores, F, Lawson, D.M, O'Connor, S.E. | | Deposit date: | 2015-08-26 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural determinants of reductive terpene cyclization in iridoid biosynthesis.
Nat.Chem.Biol., 12, 2016
|
|
7OL9
 
 | |
