2WK1
 
 | | Structure of the O-methyltransferase NovP | | Descriptor: | 1,2-ETHANEDIOL, NOVP, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Gomez Garcia, I, Stevenson, C.E.M, Uson, I, Freel Meyers, C.L, Walsh, C.T, Lawson, D.M. | | Deposit date: | 2009-06-03 | | Release date: | 2009-12-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Crystal Structure of the Novobiocin Biosynthetic Enzyme Novp: The First Representative Structure for the Tylf O-Methyltransferase Superfamily.
J.Mol.Biol., 395, 2010
|
|
5M0T
 
 | | Alpha-ketoglutarate-dependent non-heme iron oxygenase EasH | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent non-heme iron oxygenase EasH, FE (II) ION, ... | | Authors: | Jakubczyk, D, Caputi, L, Stevenson, C.E.M, Lawson, D.M, O'Connor, S.E. | | Deposit date: | 2016-10-05 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of EasH (Aspergillus japonicus) - an oxidase involved in cycloclavine biosynthesis.
Chem. Commun. (Camb.), 52, 2016
|
|
5NJF
 
 | | E. coli Microcin-processing metalloprotease TldD/E (TldD H262A mutant) with pentapeptide bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ALA-ALA-ALA-ALA-ALA, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
5NJB
 
 | | E. coli Microcin-processing metalloprotease TldD/E with actinonin bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACTINONIN, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
5NJC
 
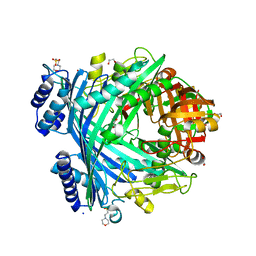 | | E. coli Microcin-processing metalloprotease TldD/E (TldD E263A mutant) with hexapeptide bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Metalloprotease PmbA, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
5NJ5
 
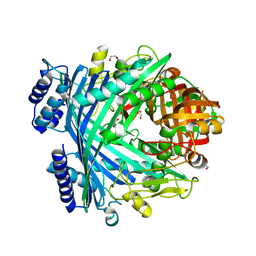 | | E. coli Microcin-processing metalloprotease TldD/E with phosphate bound | | Descriptor: | 1,2-ETHANEDIOL, Metalloprotease PmbA, Metalloprotease TldD, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
5NJ9
 
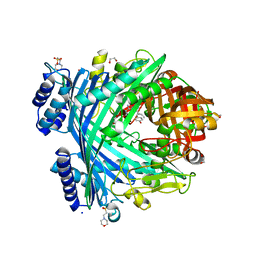 | | E. coli Microcin-processing metalloprotease TldD/E with DRVY angiotensin fragment bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ASP-ARG-VAL-TYR, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
5NJA
 
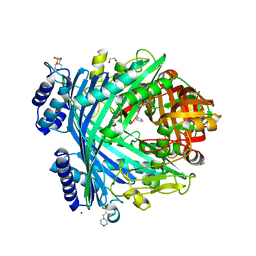 | | E. coli Microcin-processing metalloprotease TldD/E with angiotensin analogue bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HIS-PRO-PHE, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
5NZ7
 
 | | Clostridium thermocellum cellodextrin phosphorylase ligand free form | | Descriptor: | CHLORIDE ION, Cellodextrin phosphorylase | | Authors: | O'Neill, E.C, Pergolizzi, G, Stevenson, C.E.M, Lawson, D.M, Nepogodiev, S.A, Field, R.A. | | Deposit date: | 2017-05-12 | | Release date: | 2017-08-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cellodextrin phosphorylase from Ruminiclostridium thermocellum: X-ray crystal structure and substrate specificity analysis.
Carbohydr. Res., 451, 2017
|
|
5O98
 
 | | Binary complex of Catharanthus roseus Vitrosamine Synthase with NADP+ | | Descriptor: | Alcohol dehydrogenase 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Stavrinides, A.K, Tatsis, E.C, Dang, T.T, Caputi, L, Stevenson, C.E.M, Lawson, D.M, Schneider, B, O'Connor, S.E. | | Deposit date: | 2017-06-16 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of a Short-Chain Dehydrogenase from Catharanthus roseus that Produces a New Monoterpene Indole Alkaloid.
Chembiochem, 19, 2018
|
|
5NZ8
 
 | | Clostridium thermocellum cellodextrin phosphorylase with cellotetraose and phosphate bound | | Descriptor: | Cellodextrin phosphorylase, PHOSPHATE ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | O'Neill, E.C, Pergolizzi, G, Stevenson, C.E.M, Lawson, D.M, Nepogodiev, S.A, Field, R.A. | | Deposit date: | 2017-05-12 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cellodextrin phosphorylase from Ruminiclostridium thermocellum: X-ray crystal structure and substrate specificity analysis.
Carbohydr. Res., 451, 2017
|
|
6F9Q
 
 | | Binary complex of a 7S-cis-cis-nepetalactol cyclase from Nepeta mussinii with NAD+ | | Descriptor: | 7S-cis-cis-nepetalactol cyclase, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Lichman, B.R, Kamileen, M.O, Titchiner, G, Saalbach, G, Stevenson, C.E.M, Lawson, D.M, O'Connor, S.E. | | Deposit date: | 2017-12-15 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Uncoupled activation and cyclization in catmint reductive terpenoid biosynthesis.
Nat. Chem. Biol., 15, 2019
|
|
6F8J
 
 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 6-[(ethylcarbamoyl)amino]-4-(1H-pyrazol-1-yl)-N-(pyridin-3-yl)pyridine-3-carboxamide | | Descriptor: | 6-(ethylcarbamoylamino)-4-pyrazol-1-yl-~{N}-pyridin-3-yl-pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-13 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
6F96
 
 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 6-[(ethylcarbamoyl)amino]-4-[(4-methoxyphenyl)amino]-N-(pyridin-3-yl)pyridine-3-carboxamide | | Descriptor: | 6-(ethylcarbamoylamino)-4-[(4-methoxyphenyl)amino]-~{N}-pyridin-3-yl-pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-14 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
6F86
 
 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 4-(4-bromo-1H-pyrazol-1-yl)-6-[(ethylcarbamoyl)amino]-N-(pyridin-3-yl)pyridine-3-carboxamide | | Descriptor: | 4-(4-bromanylpyrazol-1-yl)-6-(ethylcarbamoylamino)-~{N}-pyridin-3-yl-pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-12 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
6F94
 
 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 6-[(ethylcarbamoyl)amino]-4-[(3-methyphenyl)amino]-N-(3-methyphenyl)pyridine-3-carboxamide | | Descriptor: | 6-(ethylcarbamoylamino)-~{N}-(3-methylphenyl)-4-[(3-methylphenyl)amino]pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
6F9H
 
 | | Crystal structure of Barley Beta-Amylase complexed with 4-S-alpha-D-glucopyranosyl-(1,4-dideoxy-4-thio-nojirimycin) | | Descriptor: | 1,4-dideoxy-4-thio-nojirimycin, Beta-amylase, CHLORIDE ION, ... | | Authors: | Moncayo, M.A, Rodrigues, L.L, Stevenson, C.E.M, Ruzanski, C, Rejzek, M, Lawson, D.M, Angulo, J, Field, R.A. | | Deposit date: | 2017-12-14 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis, biological and structural analysis of prospective glycosyl-iminosugar prodrugs: impact on germination
To be published
|
|
6F9L
 
 | | Crystal structure of Barley Beta-Amylase complexed with 3-Deoxy-3-fluoro-maltose | | Descriptor: | Beta-amylase, CHLORIDE ION, alpha-D-glucopyranose-(1-4)-3-deoxy-3-fluoro-alpha-D-glucopyranose | | Authors: | Tantanarat, K, Stevenson, C.E.M, Rejzek, M, Lawson, D.M, Field, R.A. | | Deposit date: | 2017-12-14 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of Barley Beta-Amylase complexed with 3-Deoxy-3-fluoro-maltose
To be published
|
|
6F9J
 
 | | Crystal structure of Barley Beta-Amylase complexed with 4-O-alpha-D-mannopyranosyl-(1-deoxynojirimycin) | | Descriptor: | Beta-amylase, CHLORIDE ION, alpha-D-mannopyranose-(1-4)-1-DEOXYNOJIRIMYCIN | | Authors: | Moncayo, M.A, Rodrigues, L.L, Stevenson, C.E.M, Ruzanski, C, Rejzek, M, Lawson, D.M, Angulo, J, Field, R.A. | | Deposit date: | 2017-12-14 | | Release date: | 2019-01-30 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Synthesis, biological and structural analysis of prospective glycosyl-iminosugar prodrugs: impact on germination
To be published
|
|
5CPS
 
 | | Disproportionating enzyme 1 from Arabidopsis - maltotriose soak | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase DPE1, chloroplastic/amyloplastic, ... | | Authors: | O'Neill, E.C, Stevenson, C.E.M, Tantanarat, K, Latousakis, D, Donaldson, M.I, Rejzek, M, Limpaseni, T, Smith, A.M, Field, R.A, Lawson, D.M. | | Deposit date: | 2015-07-21 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1).
J.Biol.Chem., 290, 2015
|
|
5CVS
 
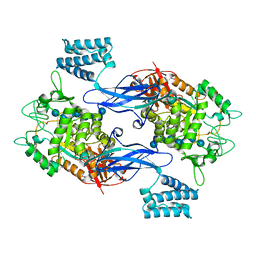 | | GlgE isoform 1 from Streptomyces coelicolor E423A mutant soaked in maltoheptaose | | Descriptor: | Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Rashid, A.M, Syson, K, Koliwer-Brandl, H, van de Weerd, R, Stevenson, C.E.M, Batey, S.F.D, Miah, F, Alber, M, Ioerger, T.R, Chandra, G, Appelmelk, B.J, Nartowski, K.P, Khimyak, Y.Z, Lawson, D.M, Jacobs, W.R, Geurtsen, J, Kalscheuer, R, Bornemann, S. | | Deposit date: | 2015-07-27 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ligand-bound structures and site-directed mutagenesis identify the acceptor and secondary binding sites of Streptomyces coelicolor maltosyltransferase GlgE.
J.Biol.Chem., 291, 2016
|
|
5DCU
 
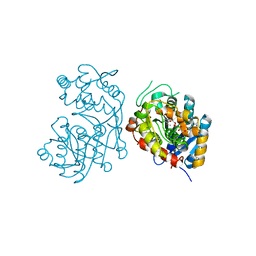 | | Iridoid synthase from Catharanthus roseus - ternary complex with NADP+ and triethylene glycol carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, Iridoid synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Caputi, L, Kries, H, Stevenson, C.E.M, Kamileen, M.O, Sherden, N.H, Geu-Flores, F, Lawson, D.M, O'Connor, S.E. | | Deposit date: | 2015-08-24 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural determinants of reductive terpene cyclization in iridoid biosynthesis.
Nat.Chem.Biol., 12, 2016
|
|
5DF1
 
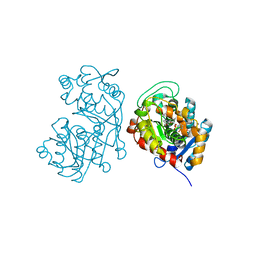 | | Iridoid synthase from Catharanthus roseus - ternary complex with NADP+ and geranic acid | | Descriptor: | (2E)-3,7-dimethylocta-2,6-dienoic acid, 1,2-ETHANEDIOL, IMIDAZOLE, ... | | Authors: | Caputi, L, Kries, H, Stevenson, C.E.M, Kamileen, M.O, Sherden, N.H, Geu-Flores, F, Lawson, D.M, O'Connor, S.E. | | Deposit date: | 2015-08-26 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural determinants of reductive terpene cyclization in iridoid biosynthesis.
Nat.Chem.Biol., 12, 2016
|
|
6GGY
 
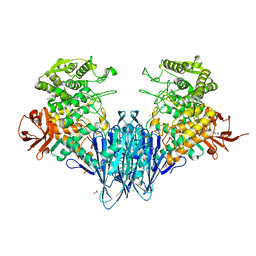 | | Paenibacillus sp. YM1 laminaribiose phosphorylase with sulphate bound | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Laminaribiose phosphorylase, ... | | Authors: | Kuhaudomlarp, S, Walpole, S, Stevenson, C.E.M, Nepogodiev, S.A, Lawson, D.M, Angulo, J, Field, R.A. | | Deposit date: | 2018-05-04 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unravelling the Specificity of Laminaribiose Phosphorylase from Paenibacillus sp. YM-1 towards Donor Substrates Glucose/Mannose 1-Phosphate by Using X-ray Crystallography and Saturation Transfer Difference NMR Spectroscopy.
Chembiochem, 20, 2019
|
|
5CQ1
 
 | | Disproportionating enzyme 1 from Arabidopsis - cycloamylose soak | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase DPE1, chloroplastic/amyloplastic, ... | | Authors: | O'Neill, E.C, Stevenson, C.E.M, Tantanarat, K, Latousakis, D, Donaldson, M.I, Rejzek, M, Limpaseni, T, Smith, A.M, Field, R.A, Lawson, D.M. | | Deposit date: | 2015-07-21 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1).
J.Biol.Chem., 290, 2015
|
|
