2K1A
 
 | |
2RMZ
 
 | |
2RN0
 
 | |
2K9J
 
 | | Integrin alphaIIb-beta3 transmembrane complex | | Descriptor: | Integrin alpha-IIb light chain, Integrin beta-3 | | Authors: | Lau, T, Kim, C, Ginsberg, M.H, Ulmer, T.S. | | Deposit date: | 2008-10-15 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the integrin alphaIIbbeta3 transmembrane complex explains integrin transmembrane signalling
Embo J., 28, 2009
|
|
1H0Z
 
 | | LEKTI domain six | | Descriptor: | SERINE PROTEASE INHIBITOR KAZAL-TYPE 5, CONTAINS HEMOFILTRATE PEPTIDE HF6478, HEMOFILTRATE PEPTIDE HF7665 | | Authors: | Lauber, T, Roesch, P, Marx, U.C. | | Deposit date: | 2002-07-01 | | Release date: | 2003-06-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Homologous Proteins with Different Folds: The Three-Dimensional Structures of Domains 1 and 6 of the Multiple Kazal-Type Inhibitor Lekti
J.Mol.Biol., 328, 2003
|
|
1HDL
 
 | | LEKTI domain one | | Descriptor: | SERINE PROTEINASE INHIBITOR LEKTI | | Authors: | Lauber, T, Roesch, P, Marx, U.C. | | Deposit date: | 2000-11-16 | | Release date: | 2001-11-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Homologous Proteins with Different Folds: The Three-Dimensional Structures of Domains 1 and 6 of the Multiple Kazal-Type Inhibitor Lekti
J.Mol.Biol., 328, 2003
|
|
6NBX
 
 | | T.elongatus NDH (data-set 2) | | Descriptor: | IRON/SULFUR CLUSTER, NAD(P)H-quinone oxidoreductase chain 4 1, NAD(P)H-quinone oxidoreductase subunit 1, ... | | Authors: | Laughlin, T.G, Bayne, A, Trempe, J.-F, Savage, D.F, Davies, K.M. | | Deposit date: | 2018-12-10 | | Release date: | 2019-02-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the complex I-like molecule NDH of oxygenic photosynthesis.
Nature, 566, 2019
|
|
6NBQ
 
 | | T.elongatus NDH (data-set 1) | | Descriptor: | IRON/SULFUR CLUSTER, NAD(P)H-quinone oxidoreductase chain 4 1, NAD(P)H-quinone oxidoreductase subunit 2, ... | | Authors: | Laughlin, T.G, Bayne, A, Trempe, J.-F, Savage, D.F, Davies, K.M. | | Deposit date: | 2018-12-09 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the complex I-like molecule NDH of oxygenic photosynthesis.
Nature, 566, 2019
|
|
6NBY
 
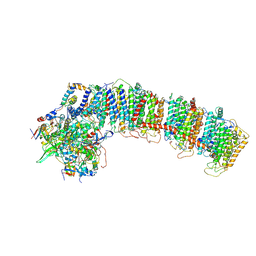 | | T.elongatus NDH (composite model) | | Descriptor: | IRON/SULFUR CLUSTER, NAD(P)H-quinone oxidoreductase chain 4 1, NAD(P)H-quinone oxidoreductase subunit 1, ... | | Authors: | Laughlin, T.G, Bayne, A, Trempe, J.-F, Savage, D.F, Davies, K.M. | | Deposit date: | 2018-12-10 | | Release date: | 2019-02-27 | | Last modified: | 2020-04-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the complex I-like molecule NDH of oxygenic photosynthesis.
Nature, 566, 2019
|
|
7SQQ
 
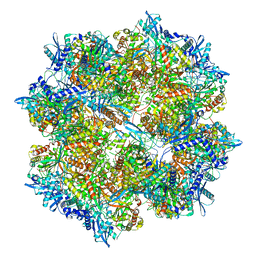 | | 201Phi2-1 Chimallin Cubic (O, 24mer) assembly | | Descriptor: | Chimallin | | Authors: | Laughlin, T.G, Deep, A, Prichard, A.M, Seitz, C, Gu, Y, Enustun, E, Suslov, S, Khanna, K, Birkholz, E.A, Amaro, R.E, Pogliano, J, Corbett, K.D, Villa, E. | | Deposit date: | 2021-11-06 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Architecture and self-assembly of the jumbo bacteriophage nuclear shell.
Nature, 608, 2022
|
|
7SQT
 
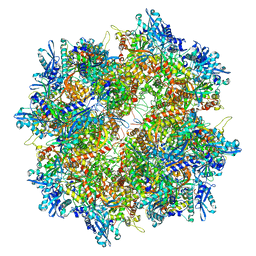 | | Goslar chimallin cubic (O, 24mer) assembly | | Descriptor: | Chimallin | | Authors: | Laughlin, T.G, Deep, A, Prichard, A.M, Seitz, C, Gu, Y, Enustun, E, Suslov, S, Khanna, K, Birkholz, E.A, Amaro, R.E, Pogliano, J, Corbett, K.D, Villa, E. | | Deposit date: | 2021-11-06 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Architecture and self-assembly of the jumbo bacteriophage nuclear shell.
Nature, 608, 2022
|
|
4I2Z
 
 | |
2R3U
 
 | | Crystal structure of the PDZ deletion mutant of DegS | | Descriptor: | Protease degS | | Authors: | Clausen, T, Kurzbauer, R. | | Deposit date: | 2007-08-30 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Regulation of the sigmaE stress response by DegS: how the PDZ domain keeps the protease inactive in the resting state and allows integration of different OMP-derived stress signals upon folding stress.
Genes Dev., 21, 2007
|
|
2R3Y
 
 | |
3H0D
 
 | |
3HGS
 
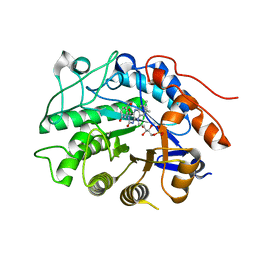 | | Crystal structure of tomato OPR3 in complex with pHB | | Descriptor: | 12-oxophytodienoate reductase 3, FLAVIN MONONUCLEOTIDE, P-HYDROXYBENZOIC ACID | | Authors: | Clausen, T, Breithaupt, C. | | Deposit date: | 2009-05-14 | | Release date: | 2009-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of substrate specificity of plant 12-oxophytodienoate reductases.
J.Mol.Biol., 392, 2009
|
|
3HGO
 
 | |
3HGR
 
 | | Crystal structure of tomato OPR1 in complex with pHB | | Descriptor: | 12-oxophytodienoate reductase 1, FLAVIN MONONUCLEOTIDE, P-HYDROXYBENZOIC ACID | | Authors: | Clausen, T, Breithaupt, C. | | Deposit date: | 2009-05-14 | | Release date: | 2009-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of substrate specificity of plant 12-oxophytodienoate reductases.
J.Mol.Biol., 392, 2009
|
|
7SQR
 
 | | 201phi2-1 Chimallin localized tetramer reconstruction | | Descriptor: | Chimallin | | Authors: | Laughlin, T.G, Deep, A, Prichard, A.M, Seitz, C, Gu, Y, Enustun, E, Suslov, S, Khanna, K, Birkholz, E.A, Amaro, R.E, Pogliano, J, Corbett, K.D, Villa, E. | | Deposit date: | 2021-11-06 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Architecture and self-assembly of the jumbo bacteriophage nuclear shell.
Nature, 608, 2022
|
|
7SQU
 
 | | Goslar chimallin C4 tetramer localized reconstruction | | Descriptor: | Chimallin | | Authors: | Laughlin, T.G, Deep, A, Prichard, A.M, Seitz, C, Gu, Y, Enustun, E, Suslov, S, Khanna, K, Birkholz, E.A, Amaro, R.E, Pogliano, J, Corbett, K.D, Villa, E. | | Deposit date: | 2021-11-06 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Architecture and self-assembly of the jumbo bacteriophage nuclear shell.
Nature, 608, 2022
|
|
7SQS
 
 | | 201phi2-1 Chimallin C1 localized reconstruction | | Descriptor: | Chimallin | | Authors: | Laughlin, T.G, Deep, A, Prichard, A.M, Seitz, C, Gu, Y, Enustun, E, Suslov, S, Khanna, K, Birkholz, E.A, Amaro, R.E, Pogliano, J, Corbett, K.D, Villa, E. | | Deposit date: | 2021-11-06 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Architecture and self-assembly of the jumbo bacteriophage nuclear shell.
Nature, 608, 2022
|
|
7SQV
 
 | | Goslar chimallin C1 localized reconstruction | | Descriptor: | Chimallin | | Authors: | Laughlin, T.G, Deep, A, Prichard, A.M, Seitz, C, Gu, Y, Enustun, E, Suslov, S, Khanna, K, Birkholz, E.A, Amaro, R.E, Pogliano, J, Corbett, K.D, Villa, E. | | Deposit date: | 2021-11-06 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Architecture and self-assembly of the jumbo bacteriophage nuclear shell.
Nature, 608, 2022
|
|
4I2W
 
 | |
3GSI
 
 | | Crystal structure of D552A dimethylglycine oxidase mutant of Arthrobacter globiformis in complex with tetrahydrofolate | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Tralau, T, Lafite, P, Levy, C, Combe, J.P, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An internal reaction chamber in dimethylglycine oxidase provides efficient protection from exposure to toxic formaldehyde.
J.Biol.Chem., 284, 2009
|
|
3QO6
 
 | | Crystal structure analysis of the plant protease Deg1 | | Descriptor: | Protease Do-like 1, chloroplastic, peptide | | Authors: | Clausen, T. | | Deposit date: | 2011-02-09 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural adaptation of the plant protease Deg1 to repair photosystem II during light exposure.
Nat.Struct.Mol.Biol., 18, 2011
|
|
