6DZ1
 
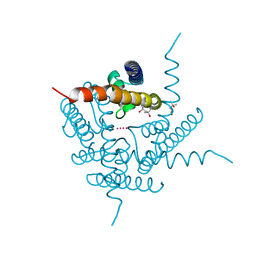 | | Studies of Ion Transport in K+ Channels | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein | | Authors: | Langan, P.S, Vandavasi, V.G, Weiss, K.L, Wagner, A, Duman, R, El Omari, K, Afonine, P.V, Coates, L. | | Deposit date: | 2018-07-02 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Anomalous X-ray diffraction studies of ion transport in K+channels.
Nat Commun, 9, 2018
|
|
6C78
 
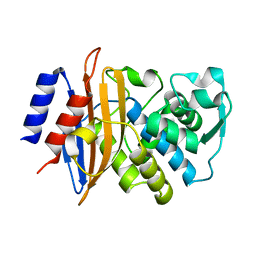 | | Substrate Binding Induces Conformational Changes In A Class A Beta Lactamase That Primes It For Catalysis | | Descriptor: | Beta-lactamase Toho-1 | | Authors: | Langan, P.S, Vandavasi, V.G, Cooper, S.J, Weiss, K.L, Ginell, S.L, Parks, J.M, Coates, L. | | Deposit date: | 2018-01-22 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | NEUTRON DIFFRACTION (1.75 Å) | | Cite: | Substrate Binding Induces Conformational Changes in a Class A Beta-lactamase That Prime It for Catalysis
Acs Catalysis, 8, 2018
|
|
6UFE
 
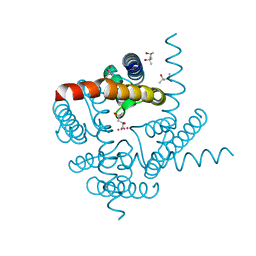 | | The structure of a potassium selective ion channel at atomic resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Transporter | | Authors: | Langan, P.S, Vandavasi, V.G, Sullivan, B, Afonine, P.V, Weiss, K.L. | | Deposit date: | 2019-09-24 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The structure of a potassium-selective ion channel reveals a hydrophobic gate regulating ion permeation.
Iucrj, 7, 2020
|
|
6U58
 
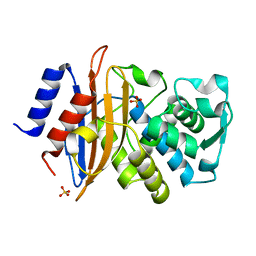 | | Toho1 Beta Lactamase Glu166Gln Mutant | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Langan, P.S, Sullivan, B, Weiss, K.L. | | Deposit date: | 2019-08-27 | | Release date: | 2020-02-19 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.9 Å), X-RAY DIFFRACTION | | Cite: | Probing the Role of the Conserved Residue Glu166 in a Class A Beta-Lactamase Using Neutron and X-ray Protein Crystallography
Acta Crystallogr.,Sect.D, 76, 2020
|
|
4HKL
 
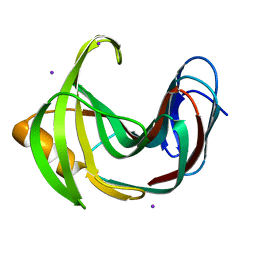 | | Crystal Structures of Mutant Endo-beta-1,4-xylanase II Complexed with substrate (1.15 A) and Products (1.6 A) | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Langan, P, Wan, Q, Coates, L, Kovalevsky, A. | | Deposit date: | 2012-10-15 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4HKO
 
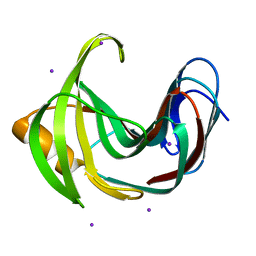 | | Crystal Structures of Mutant Endo-beta-1,4-xylanase II (E177Q) in the apo form | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Langan, P, Wan, Q, Coates, L, Kovalevsky, A. | | Deposit date: | 2012-10-15 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4HK8
 
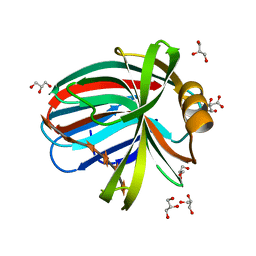 | | Crystal Structures of Mutant Endo- -1,4-xylanase II Complexed with substrate (1.15 A) and Products (1.6 A) | | Descriptor: | CITRIC ACID, Endo-1,4-beta-xylanase 2, GLYCEROL, ... | | Authors: | Langan, P, Wan, Q, Coates, L, Kovalevsky, A. | | Deposit date: | 2012-10-15 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.151 Å) | | Cite: | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4HK9
 
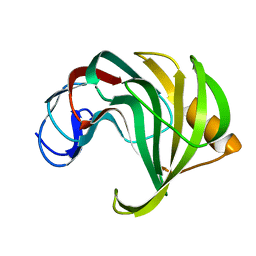 | | Crystal Structures of Mutant Endo-beta-1,4-xylanase II Complexed with substrate (1.15 A) and Products (1.6 A) | | Descriptor: | Endo-1,4-beta-xylanase 2, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Langan, P, Wan, Q, Coates, L, Kovalevsky, A. | | Deposit date: | 2012-10-15 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5EXU
 
 | |
5C6E
 
 | | Joint X-ray/neutron structure of equine cyanomet hemoglobin in R state | | Descriptor: | CYANIDE ION, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Dajnowicz, S, Sean, S, Hanson, B.L, Fisher, S.Z, Langan, P, Kovalevsky, A.Y, Mueser, T.C. | | Deposit date: | 2015-06-22 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Visualizing the Bohr effect in hemoglobin: neutron structure of equine cyanomethemoglobin in the R state and comparison with human deoxyhemoglobin in the T state.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5EBJ
 
 | | Joint X-ray/neutron structure of reversibly photoswitching chromogenic protein, Dathail | | Descriptor: | photoswitching chromogenic protein | | Authors: | Kovalevsky, A.Y, Langan, P.S, Bradbury, A.R.M. | | Deposit date: | 2015-10-19 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-15 | | Method: | NEUTRON DIFFRACTION (2.5 Å), X-RAY DIFFRACTION | | Cite: | Evolution and characterization of a new reversibly photoswitching chromogenic protein, Dathail.
J.Mol.Biol., 428, 2016
|
|
2QWS
 
 | |
4XQW
 
 | | X-ray structure analysis of xylanase-N44E with MES at pH6.0 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-20 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XPV
 
 | | Neutron and X-ray structure analysis of xylanase: N44D at pH6 | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-18 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XQ4
 
 | | X-ray structure analysis of xylanase - N44D | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-19 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XQD
 
 | | X-ray structure analysis of xylanase-WT at pH4.0 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-19 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2PKY
 
 | | The Effect of Deuteration on Protein Structure A High Resolution Comparison of Hydrogenous and Perdeuterated Haloalkane Dehalogenase | | Descriptor: | Haloalkane dehalogenase | | Authors: | Liu, X, Hanson, L, Langan, P, Viola, R.E. | | Deposit date: | 2007-04-18 | | Release date: | 2007-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The effect of deuteration on protein structure: a high-resolution comparison of hydrogenous and perdeuterated haloalkane dehalogenase.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
5EB7
 
 | |
2YXP
 
 | | The Effect of Deuteration on Protein Structure A High Resolution Comparison of Hydrogenous and Perdeuterated Haloalkane Dehalogenase | | Descriptor: | Haloalkane dehalogenase | | Authors: | Liu, X, Hanson, L, Langan, P, Viola, R.E. | | Deposit date: | 2007-04-27 | | Release date: | 2007-09-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The effect of deuteration on protein structure: a high-resolution comparison of hydrogenous and perdeuterated haloalkane dehalogenase.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3BYC
 
 | | Joint neutron and X-ray structure of diisopropyl fluorophosphatase. Deuterium occupancies are 1-Q, where Q is occupancy of H | | Descriptor: | CALCIUM ION, Diisopropyl-fluorophosphatase | | Authors: | Blum, M.-M, Mustyakimov, M, Ruterjans, H, Schoenborn, B.P, Langan, P, Chen, J.C.-H. | | Deposit date: | 2008-01-15 | | Release date: | 2009-01-27 | | Last modified: | 2024-02-21 | | Method: | NEUTRON DIFFRACTION (2.2 Å), X-RAY DIFFRACTION | | Cite: | Rapid determination of hydrogen positions and protonation states of diisopropyl fluorophosphatase by joint neutron and X-ray diffraction refinement.
Proc.Natl.Acad.Sci.Usa, 106, 2009
|
|
3QBA
 
 | | Reintroducing Electrostatics into Macromolecular Crystallographic Refinement: Z-DNA (X-ray) | | Descriptor: | Z-DNA | | Authors: | Fenn, T.D, Schnieders, M.J, Mustyakimov, M, Wu, C, Langan, P, Pande, V.S, Brunger, A.T. | | Deposit date: | 2011-01-12 | | Release date: | 2011-03-02 | | Last modified: | 2024-02-21 | | Method: | NEUTRON DIFFRACTION (1.53 Å), X-RAY DIFFRACTION | | Cite: | Reintroducing electrostatics into macromolecular crystallographic refinement: application to neutron crystallography and DNA hydration.
Structure, 19, 2011
|
|
3CWH
 
 | | D-xylose Isomerase in complex with linear product, per-deuterated xylulose | | Descriptor: | D-XYLULOSE, HYDROXIDE ION, MAGNESIUM ION, ... | | Authors: | Kovalevsky, A.Y, Langan, P, Glusker, J.P. | | Deposit date: | 2008-04-21 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | NEUTRON DIFFRACTION (2.2 Å) | | Cite: | Hydrogen location in stages of an enzyme-catalyzed reaction: time-of-flight neutron structure of D-xylose isomerase with bound D-xylulose
Biochemistry, 47, 2008
|
|
5EB6
 
 | |
5EJU
 
 | |
3X2W
 
 | | Michaelis complex of cAMP-dependent Protein Kinase Catalytic Subunit | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CARBONATE ION, MAGNESIUM ION, ... | | Authors: | Das, A, Langan, P, Gerlits, O, Kovalevsky, A.Y, Heller, W.T. | | Deposit date: | 2015-01-02 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Protein Kinase A Catalytic Subunit Primed for Action: Time-Lapse Crystallography of Michaelis Complex Formation.
Structure, 23, 2015
|
|
