1C8B
 
 | | CRYSTAL STRUCTURE OF A NOVEL GERMINATION PROTEASE FROM SPORES OF BACILLUS MEGATERIUM: STRUCTURAL REARRANGEMENTS AND ZYMOGEN ACTIVATION | | Descriptor: | SPORE PROTEASE | | Authors: | Ponnuraj, K, Rowland, S, Nessi, C, Setlow, P, Jedrzejas, M.J. | | Deposit date: | 2000-05-03 | | Release date: | 2001-05-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a novel germination protease from spores of Bacillus megaterium: structural arrangement and zymogen activation.
J.Mol.Biol., 300, 2000
|
|
6WA4
 
 | | Solution NMR structure of the unmyristoylated feline immunodeficiency virus matrix protein | | Descriptor: | Matrix protein | | Authors: | Brown, J.B, Summers, H.R, Brown, L.A, Marchant, J, Canova, P.N, O'Hern, C.T, Abbott, S.T, Nyaunu, C, Maxwell, S, Johnson, T, Moser, M.B, Carter, H, Ablan, S, Freed, E.O, Summers, M.F. | | Deposit date: | 2020-03-24 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Mechanistic Studies of the Rare Myristoylation Signal of the Feline Immunodeficiency Virus.
J.Mol.Biol., 432, 2020
|
|
6WA5
 
 | | Solution NMR Structure of the G4L/Q5K/G6S (NOS) Unmyristoylated Feline Immunodeficiency Virus Matrix Protein | | Descriptor: | Matrix protein | | Authors: | Brown, J.B, Summers, H.R, Brown, L.A, Marchant, J, Canova, P.N, O'Hern, C.T, Abbott, S.T, Nyaunu, C, Maxwell, S, Johnson, T, Moser, M.B, Ablan, S.A, Carter, H, Freed, E.O, Summers, M.F. | | Deposit date: | 2020-03-24 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Mechanistic Studies of the Rare Myristoylation Signal of the Feline Immunodeficiency Virus.
J.Mol.Biol., 432, 2020
|
|
4GBJ
 
 | |
7THW
 
 | | Crystal Structure of the Soluble Domain of the Putative OmpA -Family Membrane Protein YPO0514 from Yersinia pestis | | Descriptor: | CALCIUM ION, PHOSPHATE ION, Putative OmpA-family membrane protein | | Authors: | Kim, Y, Tesar, C, Chhor, G, Clancy, S, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-12 | | Release date: | 2022-01-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Soluble Domain of the Putative OmpA -Family Membrane Protein YPO0514 from Yersinia pestis
To Be Published
|
|
1IN8
 
 | | THERMOTOGA MARITIMA RUVB T158V | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HOLLIDAY JUNCTION DNA HELICASE RUVB | | Authors: | Putnam, C.D, Clancy, S.B, Tsuruta, H, Wetmur, J.G, Tainer, J.A. | | Deposit date: | 2001-05-12 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of the RuvB Holliday junction branch migration motor.
J.Mol.Biol., 311, 2001
|
|
5L6Q
 
 | | Refolded AL protein from cardiac amyloidosis | | Descriptor: | CARBONATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Annamalai, K, Liberta, F, Vielberg, M.-T, Lilie, H, Guehrs, K.-H, Schierhorn, A, Koehler, R, Schmidt, A, Haupt, C, Hegenbart, O, Schoenland, S, Groll, M, Faendrich, M. | | Deposit date: | 2016-05-31 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Common Fibril Structures Imply Systemically Conserved Protein Misfolding Pathways In Vivo.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
4PUE
 
 | | Extracellulr Xylanase from Geobacillus stearothermophilus: E159Q mutant, with xylotetraose in active site | | Descriptor: | CHLORIDE ION, Endo-1,4-beta-xylanase, ZINC ION, ... | | Authors: | Dann, R.D, Solomon, H.V, Lansky, S, Ben-David, A, Lavid, N, Salama, R, Shoham, Y, Shoham, G. | | Deposit date: | 2014-03-13 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Extracellulr Xylanase from Geobacillus stearothermophilus: E159Q mutant, with xylotetraose in active site.
To be Published
|
|
1IN6
 
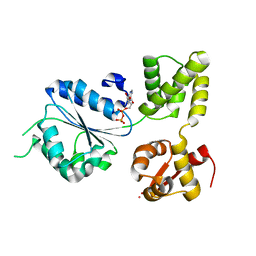 | | THERMOTOGA MARITIMA RUVB K64R MUTANT | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, COBALT (II) ION, ... | | Authors: | Putnam, C.D, Clancy, S.B, Tsuruta, H, Wetmur, J.G, Tainer, J.A. | | Deposit date: | 2001-05-12 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and mechanism of the RuvB Holliday junction branch migration motor.
J.Mol.Biol., 311, 2001
|
|
1IN7
 
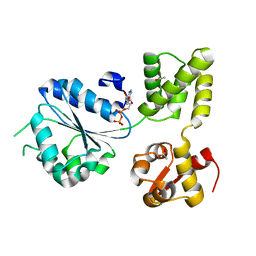 | | THERMOTOGA MARITIMA RUVB R170A | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, HOLLIDAY JUNCTION DNA HELICASE RUVB | | Authors: | Putnam, C.D, Clancy, S.B, Tsuruta, H, Wetmur, J.G, Tainer, J.A. | | Deposit date: | 2001-05-12 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of the RuvB Holliday junction branch migration motor.
J.Mol.Biol., 311, 2001
|
|
1IN5
 
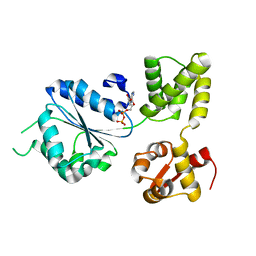 | | THERMOGOTA MARITIMA RUVB A156S MUTANT | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HOLLIDAY JUNCTION DNA HELICASE RUVB | | Authors: | Putnam, C.D, Clancy, S.B, Tsuruta, H, Gonzalez, S, Wetmur, J.G, Tainer, J.A. | | Deposit date: | 2001-05-12 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanism of the RuvB Holliday junction branch migration motor.
J.Mol.Biol., 311, 2001
|
|
2RK5
 
 | |
3RPC
 
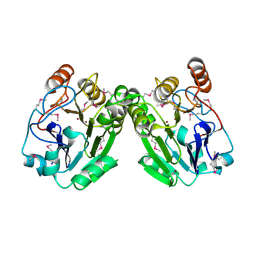 | |
3R6D
 
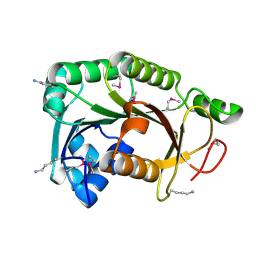 | |
4PW0
 
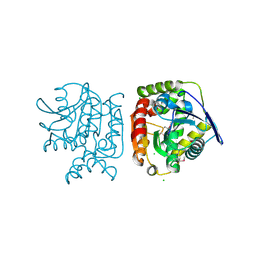 | |
4XLT
 
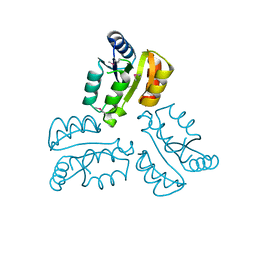 | | Crystal structure of response regulator receiver protein from Dyadobacter fermentans DSM 18053 | | Descriptor: | Response regulator receiver protein | | Authors: | Chang, C, Cuff, M, Holowicki, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-13 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of response regulator receiver protein from Dyadobacter fermentans DSM 18053
To Be Published
|
|
4HNH
 
 | | The crystal structure of a short-chain dehydrogenases/reductase (wide type) from Veillonella parvula DSM 2008 in complex with NADP | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Tan, K, Hatzos-Skintges, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-10-19 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.576 Å) | | Cite: | The crystal structure of a short-chain dehydrogenases/reductase (wide type) from Veillonella parvula DSM 2008 in complex with NADP.
To be Published
|
|
4HNG
 
 | | The crystal structure of a short-chain dehydrogenases/reductase (wide type) from Veillonella parvula DSM 2008 | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Tan, K, Hatzos-Skintges, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-10-19 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of a short-chain dehydrogenases/reductase (wide type) from Veillonella parvula DSM 2008
To be Published
|
|
4I4K
 
 | | Streptomyces globisporus C-1027 9-membered enediyne conserved protein SgcE6 | | Descriptor: | CITRIC ACID, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | Authors: | Kim, Y, Bigelow, L, Clancy, S, Babnigg, J, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-27 | | Release date: | 2012-12-12 | | Last modified: | 2016-12-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of SgcJ, an NTF2-like superfamily protein involved in biosynthesis of the nine-membered enediyne antitumor antibiotic C-1027.
J Antibiot (Tokyo), 69, 2016
|
|
4MPT
 
 | | Crystal Structure of Periplasmic binding Protein Type 1 from Bordetella pertussis Tohama I | | Descriptor: | ACETIC ACID, Putative leu/ile/val-binding protein, SODIUM ION | | Authors: | Kim, Y, Joachimiak, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-13 | | Release date: | 2013-12-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Periplasmic binding Protein Type 1 from Bordetella pertussis Tohama I
To be Published
|
|
4IAG
 
 | | Crystal structure of ZbmA, the zorbamycin binding protein from Streptomyces flavoviridis | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Zbm binding protein | | Authors: | Cuff, M.E, Bigelow, L, Bruno, C.J.P, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-12-06 | | Release date: | 2013-02-20 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Zorbamycin-Binding Protein ZbmA, the Primary Self-Resistance Element in Streptomyces flavoviridis ATCC21892.
Biochemistry, 54, 2015
|
|
5TGN
 
 | | Crystal structure of protein Sthe_2403 from Sphaerobacter thermophilus | | Descriptor: | CHLORIDE ION, GLYCEROL, Uncharacterized protein | | Authors: | Michalska, K, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-09-28 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structure of protein Sthe_2403 from Sphaerobacter thermophilus
To Be Published
|
|
5TJJ
 
 | | Crystal structure of IcIR transcriptional regulator from Alicyclobacillus acidocaldarius | | Descriptor: | GLYCEROL, Transcriptional regulator, IclR family | | Authors: | Michalska, K, Mack, J.C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-10-04 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of IcIR transcriptional regulator from Alicyclobacillus acidocaldarius
To Be Published
|
|
6GUQ
 
 | | Crystal structure of GanP, a glucose-galactose binding protein from Geobacillus stearothermophilus, in complex with glucose | | Descriptor: | Putative sugar binding protein, beta-D-glucopyranose | | Authors: | Sherf, D, Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2018-06-19 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.385 Å) | | Cite: | The crystal structure of GanP, a glucose-galactose binding protein from Geobacillus stearothermophilus, in complex with glucose
To Be Published
|
|
4LZK
 
 | |
