4OB1
 
 | | Crystal Structure of Nitrile Hydratase from Pseudonocardia thermophila bound to Butaneboronic Acid via Co-crystallization | | Descriptor: | 1-BUTANE BORONIC ACID, COBALT (II) ION, Cobalt-containing nitrile hydratase subunit alpha, ... | | Authors: | Rui, W, Salette, M, Ruslan, S, Richard, H, Dali, L. | | Deposit date: | 2014-01-06 | | Release date: | 2014-11-26 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | The active site sulfenic acid ligand in nitrile hydratases can function as a nucleophile.
J.Am.Chem.Soc., 136, 2014
|
|
5CCN
 
 | | Human Cyclophilin D Complexed with Inhibitor | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, POTASSIUM ION, ... | | Authors: | Gibson, R.P, Shore, E, Kershaw, N, Awais, M, Javed, A, Latawiec, D, Pandalaneni, S, Wen, L, Berry, N, O'Neill, P, Sutton, R, Lian, L.Y. | | Deposit date: | 2015-07-02 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human Cyclophilin D Complexed with Inhibitor
To Be Published
|
|
4O2H
 
 | | Crystal structure of BCAM1869 protein (RsaM homolog) from Burkholderia cenocepacia | | Descriptor: | protein BCAM1869 | | Authors: | Michalska, K, Chhor, G, Clancy, S, Winans, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-12-17 | | Release date: | 2014-01-22 | | Last modified: | 2014-10-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | RsaM: a transcriptional regulator of Burkholderia spp. with novel fold.
Febs J., 281, 2014
|
|
1BBU
 
 | | LYSYL-TRNA SYNTHETASE (LYSS) COMPLEXED WITH LYSINE | | Descriptor: | LYSINE, PROTEIN (LYSYL-TRNA SYNTHETASE) | | Authors: | Onesti, S, Desogus, G, Brevet, A, Chen, J, Plateau, P, Blanquet, S, Brick, P. | | Deposit date: | 1998-04-24 | | Release date: | 2000-11-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies of lysyl-tRNA synthetase: conformational changes induced by substrate binding.
Biochemistry, 39, 2000
|
|
5CCS
 
 | | Human Cyclophilin D Complexed with Inhibitor | | Descriptor: | 1-(4-aminobenzyl)-3-{2-oxo-2-[(2R)-2-phenylpyrrolidin-1-yl]ethyl}urea, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Gibson, R.P, Shore, E, Kershaw, N, Awais, M, Javed, A, Latawiec, D, Pandalaneni, S, Wen, L, Berry, N, O'Neill, P, Sutton, R, Lian, L.Y. | | Deposit date: | 2015-07-02 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human Cyclophilin D Complexed with Inhibitor
To Be Published
|
|
1BBW
 
 | | LYSYL-TRNA SYNTHETASE (LYSS) | | Descriptor: | PROTEIN (LYSYL-TRNA SYNTHETASE) | | Authors: | Onesti, S, Desogus, G, Brevet, A, Chen, J, Plateau, P, Blanquet, S, Brick, P. | | Deposit date: | 1998-04-24 | | Release date: | 2000-11-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies of lysyl-tRNA synthetase: conformational changes induced by substrate binding.
Biochemistry, 39, 2000
|
|
5CJ3
 
 | | Crystal structure of the zorbamycin binding protein (ZbmA) from Streptomyces flavoviridis with zorbamycin | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Zbm binding protein, ... | | Authors: | Chang, C, Bigelow, L, Clancy, S, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Rudolf, J.D, Ma, M, Chang, C.-Y, Lohman, J.R, Yang, D, Shen, B, Enzyme Discovery for Natural Product Biosynthesis, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-07-13 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6499 Å) | | Cite: | Crystal Structure of the Zorbamycin-Binding Protein ZbmA, the Primary Self-Resistance Element in Streptomyces flavoviridis ATCC21892.
Biochemistry, 54, 2015
|
|
5CBV
 
 | | Human Cyclophilin D Complexed with Inhibitor | | Descriptor: | FORMIC ACID, Human Cyclophilin D, POTASSIUM ION, ... | | Authors: | Gibson, R.P, Shore, E, Kershaw, N, Awais, M, Javed, A, Latawiec, D, Pandalaneni, S, Wen, L, Berry, N, O'Neill, P, Sutton, R, Lian, L.Y. | | Deposit date: | 2015-07-01 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human Cyclophilin D Complexed with Inhibitor
To Be Published
|
|
5CBW
 
 | | Human Cyclophilin D Complexed with Inhibitor. | | Descriptor: | FORMIC ACID, POTASSIUM ION, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Gibson, R.P, Shore, E, Kershaw, N, Awais, M, Javed, A, Latawiec, D, Pandalaneni, S, Wen, L, Berry, N, O'Neill, P, Sutton, R, Lian, L.Y. | | Deposit date: | 2015-07-01 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human Cyclophilin D Complexed with Inhibitor.
To Be Published
|
|
5CCQ
 
 | | Human Cyclophilin D Complexed with Inhibitor | | Descriptor: | FORMIC ACID, POTASSIUM ION, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Gibson, R.P, Shore, E, Kershaw, N, Awais, M, Javed, A, Latawiec, D, Pandalaneni, S, Wen, L, Berry, N, O'Neill, P, Sutton, R, Lian, L.Y. | | Deposit date: | 2015-07-02 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human Cyclophilin D Complexed with Inhibitor
To Be Published
|
|
5AER
 
 | | Neuronal calcium sensor-1 (NCS-1)from Rattus norvegicus complex with D2 dopamine receptor peptide from Homo sapiens | | Descriptor: | CALCIUM ION, D(2) DOPAMINE RECEPTOR, NEURONAL CALCIUM SENSOR 1, ... | | Authors: | Saleem, M, Karuppiah, V, Pandalaneni, S, Burgoyne, R, Derrick, J.P, Lian, L.Y. | | Deposit date: | 2015-01-08 | | Release date: | 2015-02-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Neuronal Calcium Sensor-1 Binds the D2 Dopamine Receptor and G-Protein-Coupled Receptor Kinase 1 (Grk1) Peptides Using Different Modes of Interactions.
J.Biol.Chem., 290, 2015
|
|
5AFP
 
 | | Neuronal calcium sensor-1 (NCS-1)from Rattus norvegicus complex with rhodopsin kinase peptide from Homo sapiens | | Descriptor: | CALCIUM ION, NEURONAL CALCIUM SENSOR 1, RHODOPSIN KINASE, ... | | Authors: | Saleem, M, Karuppiah, V, Pandalaneni, S, Burgoyne, R, Derrick, J.P, Lian, L.Y. | | Deposit date: | 2015-01-23 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Neuronal Calcium Sensor-1 Binds the D2 Dopamine Receptor and G-Protein Coupled Receptor Kinase 1 (Grk1) Peptides Using Different Modes of Interactions.
J.Biol.Chem., 290, 2015
|
|
1S1E
 
 | | Crystal Structure of Kv Channel-interacting protein 1 (KChIP-1) | | Descriptor: | CALCIUM ION, Kv channel interacting protein 1 | | Authors: | Scannevin, R.H, Wang, K.-W, Jow, F, Megules, J, Kopsco, D.C, Edris, W, Carroll, K.C, Lu, Q, Xu, W.-X, Xu, Z.-B, Katz, A.H, Olland, S, Lin, L, Taylor, M, Stahl, M, Malakian, K, Somers, W, Mosyak, L, Bowlby, M.R, Chanda, P, Rhodes, K.J. | | Deposit date: | 2004-01-06 | | Release date: | 2005-01-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Two N-terminal domains of Kv4 K(+) channels regulate binding to and modulation by KChIP1.
Neuron, 41, 2004
|
|
4N3P
 
 | | Crystal Structure of De Novo designed Serine Hydrolase OSH18, Northeast Structural Genomics Consortium (NESG) Target OR396 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Mao, L, Xiao, R, Kogan, S, Maglaqui, M, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-10-07 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal Structure of De Novo designed Serine Hydrolase OSH18, Northeast Structural Genomics Consortium (NESG) Target OR396
To be Published
|
|
4H7N
 
 | |
1RQG
 
 | | Methionyl-tRNA synthetase from Pyrococcus abyssi | | Descriptor: | Methionyl-tRNA synthetase, ZINC ION | | Authors: | Crepin, T, Schmitt, E, Blanquet, S, Mechulam, Y. | | Deposit date: | 2003-12-05 | | Release date: | 2004-05-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Three-dimensional structure of methionyl-tRNA synthetase from Pyrococcus abyssi.
Biochemistry, 43, 2004
|
|
4PS2
 
 | | Structure of the C-terminal fragment (87-165) of E.coli EAEC TssB molecule | | Descriptor: | CHLORIDE ION, Putative type VI secretion protein, ZINC ION | | Authors: | Douzi, B, Logger, L, Spinelli, S, Blangy, S, Cambillau, C, Cascales, E. | | Deposit date: | 2014-03-06 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping the tube-sheath interface within the Type VI secretion system tail
To be Published
|
|
5DFA
 
 | | 3D structure of the E323A catalytic mutant of Gan42B, a GH42 beta-galactosidase from G. stearothermophilus | | Descriptor: | Beta-galactosidase, GLYCEROL, ZINC ION | | Authors: | Solomon, H.V, Tabachnikov, O, Lansky, S, Feinberg, H, Govada, L, Chayen, N.E, Shoham, Y, Shoham, G. | | Deposit date: | 2015-08-26 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-function relationships in Gan42B, an intracellular GH42 beta-galactosidase from Geobacillus stearothermophilus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4OB2
 
 | | Crystal Structure of Nitrile Hydratase from Pseudonocardia thermophila bound to Butaneboronic Acid via Crystal Soaking | | Descriptor: | 1-BUTANE BORONIC ACID, COBALT (II) ION, Cobalt-containing nitrile hydratase subunit alpha, ... | | Authors: | Rui, W, Salette, M, Ruslan, S, Richard, H, Dali, L. | | Deposit date: | 2014-01-06 | | Release date: | 2014-11-26 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The active site sulfenic acid ligand in nitrile hydratases can function as a nucleophile.
J.Am.Chem.Soc., 136, 2014
|
|
4PSC
 
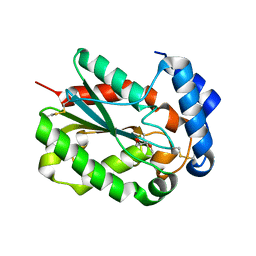 | | Structure of cutinase from Trichoderma reesei in its native form. | | Descriptor: | Carbohydrate esterase family 5, GLYCEROL | | Authors: | Roussel, A, Amara, S, Nyyssola, A, Mateos-Diaz, E, Blangy, S, Kontkanen, H, Westerholm-Parvinen, A, Carriere, F, Cambillau, C. | | Deposit date: | 2014-03-07 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | A Cutinase from Trichoderma reesei with a Lid-Covered Active Site and Kinetic Properties of True Lipases.
J.Mol.Biol., 426, 2014
|
|
1MNB
 
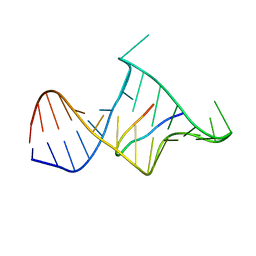 | | BIV TAT PEPTIDE (RESIDUES 68-81), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BIV TAR RNA, BIV TAT PEPTIDE | | Authors: | Puglisi, J.D, Chen, L, Blanchard, S, Frankel, A.D. | | Deposit date: | 1996-07-25 | | Release date: | 1997-01-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a bovine immunodeficiency virus Tat-TAR peptide-RNA complex.
Science, 270, 1995
|
|
4OB0
 
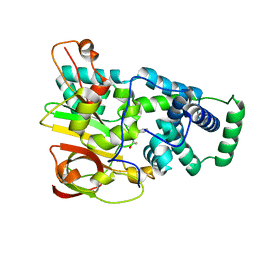 | | Crystal Structure of Nitrile Hydratase from Pseudonocardia thermophila bound to Phenyl Boronic Acid | | Descriptor: | COBALT (II) ION, Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta, ... | | Authors: | Rui, W, Salette, M, Ruslan, S, Richard, H, Dali, L. | | Deposit date: | 2014-01-06 | | Release date: | 2014-11-26 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The active site sulfenic acid ligand in nitrile hydratases can function as a nucleophile.
J.Am.Chem.Soc., 136, 2014
|
|
4OB3
 
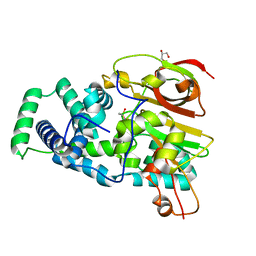 | | Crystal Structure of Nitrile Hydratase from Pseudonocardia thermophila : A Reference Structure to Boronic Acid Inhibition of Nitrile Hydratase | | Descriptor: | COBALT (II) ION, Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta, ... | | Authors: | Rui, W, Salette, M, Ruslan, S, Richard, H, Dali, L. | | Deposit date: | 2014-01-06 | | Release date: | 2014-11-26 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The active site sulfenic acid ligand in nitrile hydratases can function as a nucleophile.
J.Am.Chem.Soc., 136, 2014
|
|
4HES
 
 | | Structure of a Beta-Lactamase Class A-like Protein from Veillonella parvula. | | Descriptor: | Beta-lactamase class A-like protein, FORMIC ACID, GLYCEROL, ... | | Authors: | Cuff, M.E, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-10-04 | | Release date: | 2012-10-17 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Beta-Lactamase Class A-like Protein from Veillonella parvula.
TO BE PUBLISHED
|
|
1BJ8
 
 | | THIRD N-TERMINAL DOMAIN OF GP130, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GP130 | | Authors: | Kernebeck, T, Pflanz, S, Muller-Newen, G, Kurapkat, G, Scheek, R.M, Dijkstra, K, Heinrich, P.C, Wollmer, A, Grzesiek, S, Grotzinger, J. | | Deposit date: | 1998-07-02 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The signal transducer gp130: solution structure of the carboxy-terminal domain of the cytokine receptor homology region.
Protein Sci., 8, 1999
|
|
