2NAC
 
 | | HIGH RESOLUTION STRUCTURES OF HOLO AND APO FORMATE DEHYDROGENASE | | Descriptor: | NAD-DEPENDENT FORMATE DEHYDROGENASE, SULFATE ION | | Authors: | Lamzin, V.S, Dauter, Z, Popov, V.O, Harutyunyan, E.H, Wilson, K.S. | | Deposit date: | 1994-07-06 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structures of holo and apo formate dehydrogenase.
J.Mol.Biol., 236, 1994
|
|
2NAD
 
 | | HIGH RESOLUTION STRUCTURES OF HOLO AND APO FORMATE DEHYDROGENASE | | Descriptor: | AZIDE ION, NAD-DEPENDENT FORMATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Lamzin, V.S, Dauter, Z, Popov, V.O, Harutyunyan, E.H, Wilson, K.S. | | Deposit date: | 1994-07-06 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | High resolution structures of holo and apo formate dehydrogenase.
J.Mol.Biol., 236, 1994
|
|
1EGP
 
 | |
1YPH
 
 | | High resolution structure of bovine alpha-chymotrypsin | | Descriptor: | CHYMOTRYPSIN A, chain A, chain B, ... | | Authors: | Razeto, A, Galunsky, B, Kasche, V, Wilson, K.S, Lamzin, V.S. | | Deposit date: | 2005-01-31 | | Release date: | 2006-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | High resolution structure of native bovine alpha-chymotrypsin
To be Published
|
|
7QLP
 
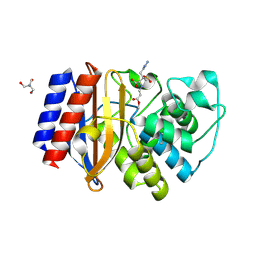 | | Structure of beta-lactamase TEM-171 complexed with tazobactam intermediate at 2.3 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Hakanpaa, J, Petrova, T, Samygina, V.R, Lamzin, V.S, Egorov, A.M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the molecular class A beta-lactamase TEM-171 and its complexes with tazobactam.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QNK
 
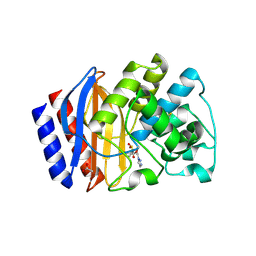 | | Structure of beta-lactamase TEM-171 complexed with tazobactam intermediate at 2.5 A resolution | | Descriptor: | ACETATE ION, Beta-lactamase TEM, TAZOBACTAM INTERMEDIATE | | Authors: | Hakanpaa, J, Petrova, T, Samygina, V.R, Lamzin, V.S, Egorov, A.M. | | Deposit date: | 2021-12-21 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the molecular class A beta-lactamase TEM-171 and its complexes with tazobactam.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QOR
 
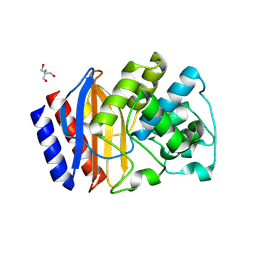 | | Structure of beta-lactamase TEM-171 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Hakanpaa, J, Petrova, T, Samygina, V.R, Chojnowski, G, Lamzin, V, Egorov, A.M. | | Deposit date: | 2021-12-28 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Crystal structures of the molecular class A beta-lactamase TEM-171 and its complexes with tazobactam.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
5LDU
 
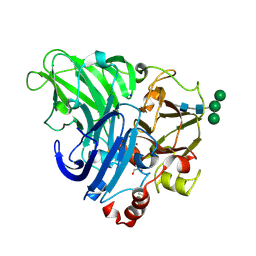 | | Recombinant high-redox potential laccase from Basidiomycete Trametes hirsuta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, GLYCEROL, ... | | Authors: | Polyakov, K.M, Fedorova, T.V, Loginov, D.S, Savinova, O.S, Lamzin, V.S, Koroleva, O.V. | | Deposit date: | 2016-06-27 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | First structure of recombinant high-redox potential laccase from Basidiomycete Trametes hirsuta.
To Be Published
|
|
2ZO5
 
 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in a complex with azide | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AZIDE ION, ... | | Authors: | Polyakov, K.M, Boyko, K.M, Slutsky, A, Tikhonova, T.V, Antipov, A.N, Zvyagilskaya, R.A, Popov, A.N, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2008-05-05 | | Release date: | 2009-05-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution structural analysis of a novel octaheme cytochrome c nitrite reductase from the haloalkaliphilic bacterium Thioalkalivibrio nitratireducens
J.Mol.Biol., 389, 2009
|
|
4LZT
 
 | | ATOMIC RESOLUTION REFINEMENT OF TRICLINIC HEW LYSOZYME AT 295K | | Descriptor: | LYSOZYME, NITRATE ION | | Authors: | Walsh, M.A, Schneider, T, Sieker, L.C, Dauter, Z, Lamzin, V, Wilson, K.S. | | Deposit date: | 1997-03-31 | | Release date: | 1998-04-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Refinement of triclinic hen egg-white lysozyme at atomic resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
3V9C
 
 | | Type-2 Cu-depleted fungus laccase from Trametes hirsuta at low dose of ionization radiation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Polyakov, K.M, Fedorova, T.V, Kurzeev, S.A, Popov, A.N, Lamzin, V.S, Koroleva, O.V. | | Deposit date: | 2011-12-26 | | Release date: | 2012-02-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of T2-depleted fungal laccase from Trametes hirsuta 072 at low and high dose of ionization radiation
To be Published
|
|
2V8Z
 
 | | Crystal Structure of YagE, a prophage protein belonging to the dihydrodipicolinic acid synthase family from E. coli K12 | | Descriptor: | YAGE | | Authors: | Manicka, S, Peleg, Y, Unger, T, Albeck, S, Dym, O, Greenblatt, H.M, Bourenkov, G, Lamzin, V, Krishnaswamy, S, Sussman, J.L. | | Deposit date: | 2007-08-16 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Yage, a Putative Dhdps Like Protein from Escherichia Coli K12.
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
3ZL4
 
 | | Antibody structural organization: Role of kappa - lambda chain constant domain switch in catalytic functionality | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, A17 ANTIBODY FAB FRAGMENT HEAVY CHAIN, A17 ANTIBODY FAB FRAGMENT LAMBDA LIGHT CHAIN | | Authors: | Chatziefthimiou, S.D, Ponomarenko, N.A, Kurkova, I.N, Smirnov, A.V, Smirnov, I.V, Lamzin, V.S, Gabibov, A.G, Wilmanns, M. | | Deposit date: | 2013-01-28 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Role of Kappa>Lambda Light-Chain Constant-Domain Switch in the Structure and Functionality of A17 Reactibody
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2V9D
 
 | | Crystal Structure of YagE, a prophage protein belonging to the dihydrodipicolinic acid synthase family from E. coli K12 | | Descriptor: | YAGE | | Authors: | Manicka, S, Peleg, Y, Unger, T, Albeck, S, Dym, O, Greenblatt, H.M, Bourenkov, G, Lamzin, V, Krishnaswamy, S, Sussman, J.L. | | Deposit date: | 2007-08-23 | | Release date: | 2008-03-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Yage, a Putative Dhdps Like Protein from Escherichia Coli K12.
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
3PXL
 
 | | Type-2 Cu-depleted fungus laccase from Trametes hirsuta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Polyakov, K.M, Fedorova, T.V, Kurzeev, S.A, Popov, A.N, Lamzin, V.S, Koroleva, O.V. | | Deposit date: | 2010-12-10 | | Release date: | 2011-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of T2-depleted fungal laccase from Trametes hirsuta at low and high dose of ionization radiation.
To be Published
|
|
2OT4
 
 | | Structure of a hexameric multiheme c nitrite reductase from the extremophile bacterium Thiolkalivibrio nitratireducens | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Polyakov, K.M, Boyko, K.M, Slutsky, A, Tikhonova, T.V, Antipov, A.N, Zvyagilskaya, R.A, Popov, A.N, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2007-02-07 | | Release date: | 2008-04-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution structural analysis of a novel octaheme cytochrome c nitrite reductase from the haloalkaliphilic bacterium Thioalkalivibrio nitratireducens.
J.Mol.Biol., 389, 2009
|
|
3NIR
 
 | | Crystal structure of small protein crambin at 0.48 A resolution | | Descriptor: | Crambin, ETHANOL | | Authors: | Schmidt, A, Teeter, M, Weckert, E, Lamzin, V.S. | | Deposit date: | 2010-06-16 | | Release date: | 2011-05-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (0.48 Å) | | Cite: | Crystal structure of small protein crambin at 0.48 A resolution
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3FPX
 
 | | Native fungus laccase from Trametes hirsuta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Polyakov, K.M, Fedorova, T.V, Stepanova, E.V, Cherkashin, E.A, Kurzeev, S.A, Strokopytov, B.V, Lamzin, V.S, Koroleva, O.V. | | Deposit date: | 2009-01-06 | | Release date: | 2009-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of native laccase from Coriolus hirsutus at 1.8 A resolution
To be Published
|
|
3D1I
 
 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in a complex with nitrite | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Polyakov, K.M, Boyko, K.M, Slutsky, A, Tikhonova, T.V, Antipov, A.N, Zvyagilskaya, R.A, Popov, A.N, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2008-05-06 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structural analysis of a novel octaheme cytochrome c nitrite reductase from the haloalkaliphilic bacterium Thioalkalivibrio nitratireducens
J.Mol.Biol., 389, 2009
|
|
5NPL
 
 | | Crystal structure of hexameric CBS-CP12 protein from bloom-forming cyanobacteria, Yb-derivative at 2.8 A resolution | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, Similar to tr|Q8YYT1|Q8YYT1, YTTERBIUM (III) ION | | Authors: | Hackenberg, C, Hakanpaa, J, Antonyuk, S.V, Dittmann, E, Lamzin, V.S. | | Deposit date: | 2017-04-17 | | Release date: | 2018-05-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural and functional insights into the unique CBS-CP12 fusion protein family in cyanobacteria.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5NMU
 
 | | Structure of hexameric CBS-CP12 protein from bloom-forming cyanobacteria | | Descriptor: | CBS-CP12, CHLORIDE ION | | Authors: | Hackenberg, C, Hakanpaa, J, Antonyuk, S.V, Dittmann, E, Lamzin, V.S. | | Deposit date: | 2017-04-07 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and functional insights into the unique CBS-CP12 fusion protein family in cyanobacteria.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5NVD
 
 | | Crystal structure of hexameric CBS-CP12 protein from bloom-forming cyanobacteria at 2.5 A resolution in P6322 crystal form | | Descriptor: | CBS-CP12 | | Authors: | Hackenberg, C, Hakanpaa, J, Eigner, C, Antonyuk, S.V, Dittmann, E, Lamzin, V.S. | | Deposit date: | 2017-05-04 | | Release date: | 2018-05-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional insights into the unique CBS-CP12 fusion protein family in cyanobacteria.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2CNQ
 
 | | Atomic resolution structure of SAICAR-synthase from Saccharomyces cerevisiae complexed with ADP, AICAR, succinate | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, ... | | Authors: | Urusova, D.V, Antonyuk, S.V, Grebenko, A.I, Levdikov, V.M, Barynin, V.V, Popov, A.N, Lamzin, V.S, Melik-Adamyan, W.R. | | Deposit date: | 2006-05-23 | | Release date: | 2006-06-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Saicar Synthase: Substrate Recognition, Conformational Flexibility and Catalysis.
To be Published
|
|
2CNV
 
 | | SAICAR-synthase from Saccharomyces cerevisiae complexed SAICAR | | Descriptor: | ASPARTIC ACID, N-{[5-AMINO-1-(5-O-PHOSPHONO-BETA-D-ARABINOFURANOSYL)-1H-IMIDAZOL-4-YL]CARBONYL}-L-ASPARTIC ACID, PHOSPHORIBOSYLAMINOIMIDAZOLE-SUCCINOCARBOXAMIDE SYNTHASE, ... | | Authors: | Urusova, D.V, Antonyuk, S.V, Grebenko, A.I, Levdikov, V.M, Barynin, V.V, Popov, A.N, Lamzin, V.S, Melik-Adamyan, V.R. | | Deposit date: | 2006-05-24 | | Release date: | 2006-06-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray Diffraction Study of the Complex of the Enzyme Saicar Synthase with the Reaction Product
Crystallogr.Rep.(Transl. Kristallografiya), 51, 2006
|
|
2CNU
 
 | | Atomic resolution structure of SAICAR-synthase from Saccharomyces cerevisiae complexed with aspartic acid | | Descriptor: | ASPARTIC ACID, PHOSPHORIBOSYLAMINOIMIDAZOLE-SUCCINOCARBOXAMIDE SYNTHASE, SULFATE ION | | Authors: | Urusova, D.V, Antonyuk, S.V, Grebenko, A.I, Levdikov, V.M, Barynin, V.V, Popov, A.N, Lamzin, V.S, Melik-Adamyan, W.R. | | Deposit date: | 2006-05-24 | | Release date: | 2006-06-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Saicar Synthase: Substrate Recognition, Conformational Flexibility and Catalysis.
To be Published
|
|
