1BUF
 
 | |
2W4P
 
 | | Human common-type acylphosphatase variant, A99G | | Descriptor: | ACYLPHOSPHATASE-1, GLYCEROL | | Authors: | Lam, S.Y, Sze, K.H, Wong, K.B. | | Deposit date: | 2008-11-29 | | Release date: | 2009-12-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Rigidifying Salt-Bridge Favors the Activity of Thermophilic Enzyme at High Temperatures at the Expense of Low-Temperature Activity.
Plos Biol., 9, 2011
|
|
2W4C
 
 | | Human common-type acylphosphatase variant, A99 | | Descriptor: | ACYLPHOSPHATASE-1 | | Authors: | Lam, S.Y, Wong, K.B. | | Deposit date: | 2008-11-25 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A Rigidifying Salt-Bridge Favors the Activity of Thermophilic Enzyme at High Temperatures at the Expense of Low-Temperature Activity.
Plos Biol., 9, 2011
|
|
1UQD
 
 | |
1UQC
 
 | |
1UQE
 
 | |
1UQG
 
 | |
1UQF
 
 | |
1UQA
 
 | |
1UQB
 
 | |
6IY5
 
 | | NMR solution structures of 5'-ATTCTATTCT-3 | | Descriptor: | DNA (5'-D(*AP*TP*TP*CP*TP*AP*TP*TP*CP*T)-3'), SODIUM ION | | Authors: | Lam, S.L, Guo, P. | | Deposit date: | 2018-12-13 | | Release date: | 2020-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Minidumbbell structures formed by ATTCT pentanucleotide repeats in spinocerebellar ataxia type 10.
Nucleic Acids Res., 48, 2020
|
|
2W4D
 
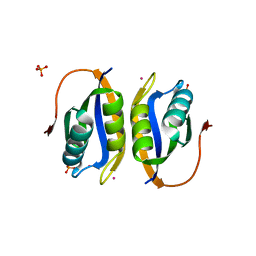 | | Acylphosphatase variant G91A from Pyrococcus horikoshii | | Descriptor: | ACYLPHOSPHATASE, PHOSPHATE ION, POTASSIUM ION | | Authors: | Lam, S.Y, Wong, K.B. | | Deposit date: | 2008-11-25 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Rigidifying Salt-Bridge Favors the Activity of Thermophilic Enzyme at High Temperatures at the Expense of Low-Temperature Activity.
Plos Biol., 9, 2011
|
|
6J37
 
 | | DNA minidumbbell structure of two CTTG repeats | | Descriptor: | DNA (5'-D(*CP*TP*TP*GP*CP*TP*TP*G)-3'), SODIUM ION | | Authors: | Lam, S.L, Guo, P. | | Deposit date: | 2019-01-04 | | Release date: | 2019-05-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Unprecedented hydrophobic stabilizations from a reverse wobble T·T mispair in DNA minidumbbell.
J.Biomol.Struct.Dyn., 38, 2020
|
|
1K2J
 
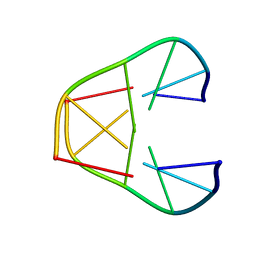 | | NMR MINIMIZED AVERAGE STRUCTURE OF d(CGTACG)2 | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3' | | Authors: | Lam, S.L, Ip, L.N. | | Deposit date: | 2001-09-27 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Low temperature solution structures and base pair stacking of double helical d(CGTACG)(2).
J.Biomol.Struct.Dyn., 19, 2002
|
|
1K2K
 
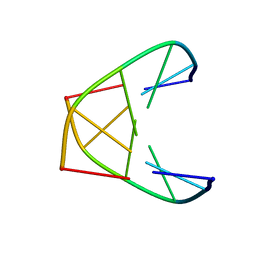 | | NMR MINIMIZED AVERAGE STRUCTURE OF d(CGTACG)2 | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3' | | Authors: | Lam, S.L, Ip, L.N. | | Deposit date: | 2001-09-28 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Low temperature solution structures and base pair stacking of double helical d(CGTACG)(2).
J.Biomol.Struct.Dyn., 19, 2002
|
|
7LP4
 
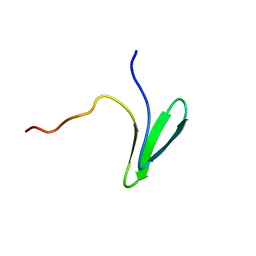 | | Structure of Nedd4L WW3 domain | | Descriptor: | E3 ubiquitin-protein ligase NEDD4-like | | Authors: | Alam, S.L, Alian, A, Thompson, T, Rheinemann, L, Volkman, B.F, Peterson, F.C, Sundquist, W.I. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Interactions between AMOT PPxY motifs and NEDD4L WW domains function in HIV-1 release.
J.Biol.Chem., 297, 2021
|
|
7LP5
 
 | | Structure of Nedd4L WW3 domain | | Descriptor: | Angiomotin,E3 ubiquitin-protein ligase NEDD4-like | | Authors: | Alam, S.L, Alian, A, Thompson, T, Rheinemann, L, Volkman, B.F, Peterson, F.C, Sundquist, W.I. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Interactions between AMOT PPxY motifs and NEDD4L WW domains function in HIV-1 release.
J.Biol.Chem., 297, 2021
|
|
3D97
 
 | |
2P1H
 
 | | Rapid Folding and Unfolding of Apaf-1 CARD | | Descriptor: | Apoptotic protease-activating factor 1, ZINC ION | | Authors: | Milam, S.L, Nicely, N.I, Feeney, B, Mattos, C, Clark, A.C. | | Deposit date: | 2007-03-05 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Rapid Folding and Unfolding of Apaf-1 CARD.
J.Mol.Biol., 369, 2007
|
|
3D95
 
 | |
3D96
 
 | |
3CWK
 
 | |
1FJP
 
 | | NMR Solution Structure of Phospholamban (C41F) | | Descriptor: | CARDIAC PHOSPHOLAMBAN | | Authors: | Lamberth, S, Griesinger, C, Schmid, H, Carafoli, E, Muenchbach, M, Vorherr, T, Krebs, J. | | Deposit date: | 2000-08-08 | | Release date: | 2000-09-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Phospholamban
HELV.CHIM.ACTA, 83, 2000
|
|
1FJK
 
 | | NMR Solution Structure of Phospholamban (C41F) | | Descriptor: | CARDIAC PHOSPHOLAMBAN | | Authors: | Lamberth, S, Griesinger, C, Schmid, H, Carafoli, E, Muenchbach, M, Vorherr, T, Krebs, J. | | Deposit date: | 2000-08-08 | | Release date: | 2000-09-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Phospholamban
HELV.CHIM.ACTA, 83, 2000
|
|
2GQB
 
 | | Solution Structure of a conserved unknown protein RPA2825 from Rhodopseudomonas palustris; (Northeast Structural Genomics Consortium Target RpT4; Ontario Centre for Structural Proteomics Target rp2812 ) | | Descriptor: | conserved hypothetical protein | | Authors: | Srisailam, S, Lukin, J.A, Yee, A, Lemak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-04-20 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a conserved unknown protein RPA2825 from Rhodopseudomonas palustris
To be Published
|
|
